|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
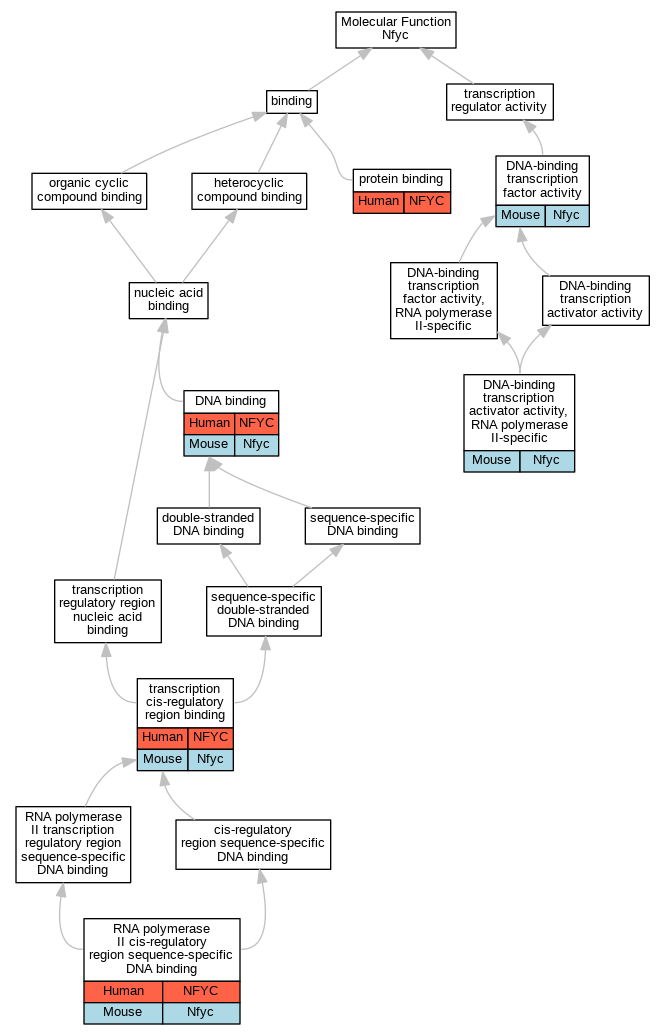
| Molecular Function | GO:0003677 | DNA binding | NFYC | IDA | Human | PMID:15243141 | |
| Molecular Function | GO:0003677 | DNA binding | Nfyc | IDA | Mouse | PMID:15601870 | |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | Nfyc | IDA | Mouse | MGI:MGI:3525928|PMID:15601870 | |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | Nfyc | IDA | Mouse | PMID:15601870 | |
| Molecular Function | GO:0005515 | protein binding | NFYC | IPI | UniProtKB:P25208 | Human | PMID:12401788 |
| Molecular Function | GO:0005515 | protein binding | NFYC | IPI | UniProtKB:P23511 | Human | PMID:15243141 |
| Molecular Function | GO:0005515 | protein binding | NFYC | IPI | UniProtKB:P23511 | Human | PMID:20936779 |
| Molecular Function | GO:0005515 | protein binding | NFYC | IPI | UniProtKB:P25208 | Human | PMID:20936779 |
| Molecular Function | GO:0005515 | protein binding | NFYC | IPI | UniProtKB:P25208 | Human | PMID:21988832 |
| Molecular Function | GO:0005515 | protein binding | NFYC | IPI | UniProtKB:P23511 | Human | PMID:23332751 |
| Molecular Function | GO:0005515 | protein binding | NFYC | IPI | UniProtKB:P25208 | Human | PMID:26496610 |
| Molecular Function | GO:0005515 | protein binding | NFYC | IPI | UniProtKB:P23511 | Human | PMID:35140242 |
| Molecular Function | GO:0005515 | protein binding | NFYC | IPI | UniProtKB:P25208 | Human | PMID:35140242 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | NFYC | IDA | Human | PMID:11256944 | |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | Nfyc | IDA | Mouse | MGI:MGI:3525928|PMID:15601870 | |
| Molecular Function | GO:0000976 | transcription cis-regulatory region binding | NFYC | IDA | Human | PMID:11256944 | |
| Molecular Function | GO:0000976 | transcription cis-regulatory region binding | Nfyc | IDA | Mouse | PMID:15516209 | |
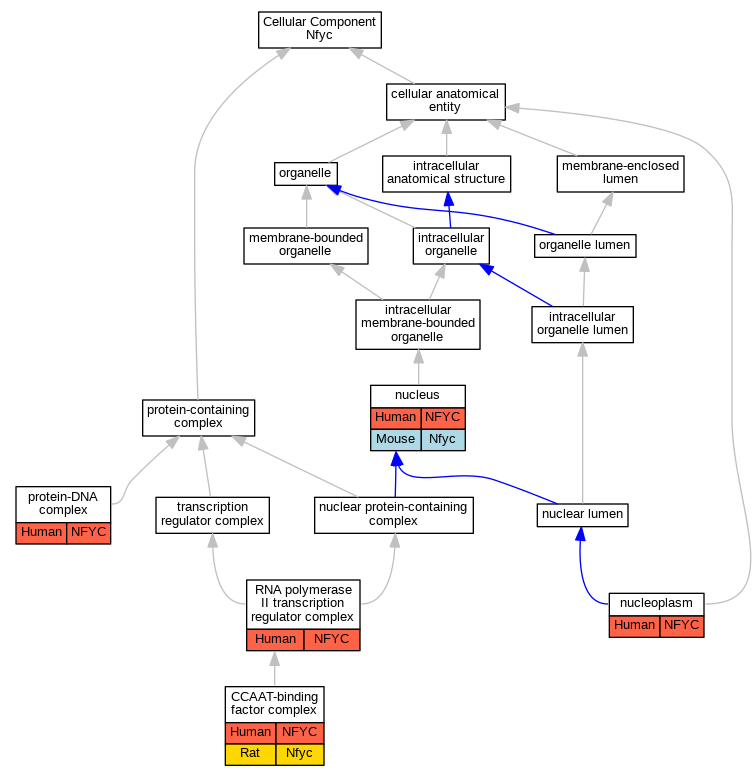
| Cellular Component | GO:0016602 | CCAAT-binding factor complex | NFYC | IPI | Human | PMID:23332751 | |
| Cellular Component | GO:0016602 | CCAAT-binding factor complex | NFYC | IDA | Human | PMID:15243141 | |
| Cellular Component | GO:0016602 | CCAAT-binding factor complex | Nfyc | IDA | Rat | PMID:7878029|RGD:729287 | |
| Cellular Component | GO:0005654 | nucleoplasm | NFYC | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005634 | nucleus | NFYC | IDA | Human | PMID:15243141 | |
| Cellular Component | GO:0005634 | nucleus | NFYC | IDA | Human | PMID:15964792 | |
| Cellular Component | GO:0005634 | nucleus | Nfyc | IDA | Mouse | PMID:19734906 | |
| Cellular Component | GO:0032993 | protein-DNA complex | NFYC | IDA | Human | PMID:11256944 | |
| Cellular Component | GO:0090575 | RNA polymerase II transcription regulator complex | NFYC | IDA | Human | PMID:11256944 | |
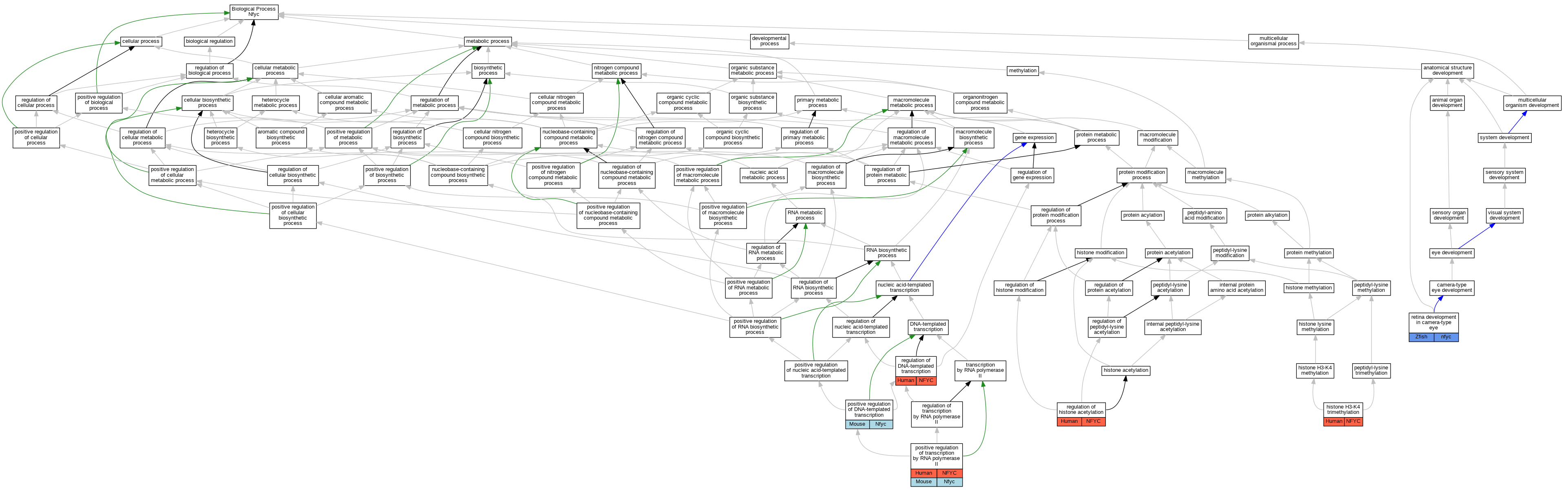
| Biological Process | GO:0080182 | histone H3-K4 trimethylation | NFYC | IDA | Human | PMID:21445285 | |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | Nfyc | IDA | Mouse | PMID:15601870 | |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | NFYC | IDA | Human | PMID:23332751 | |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Nfyc | IDA | Mouse | MGI:MGI:3525928|PMID:15601870 | |
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | NFYC | IDA | Human | PMID:15243141 | |
| Biological Process | GO:0035065 | regulation of histone acetylation | NFYC | IDA | Human | PMID:21304275 | |
| Biological Process | GO:0060041 | retina development in camera-type eye | nfyc | IMP | ZFIN:ZDB-GENO-040720-1 | Zfish | PMID:15716491 |
| Biological Process | GO:0060041 | retina development in camera-type eye | nfyc | IMP | ZFIN:ZDB-GENO-040720-2 | Zfish | PMID:15716491 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 07/05/2024 MGI 6.24 |

|
|
|
||