| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
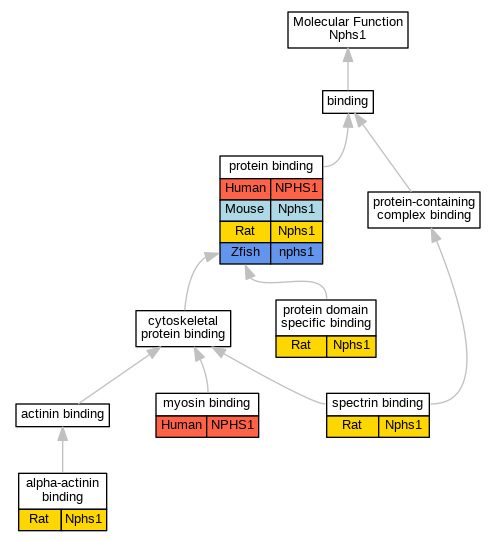
| Molecular Function | GO:0051393 | alpha-actinin binding | Nphs1 | IPI | RGD:61816 | Rat | PMID:15331416|RGD:1600864 |
| Molecular Function | GO:0017022 | myosin binding | NPHS1 | IPI | UniProtKB:O00159 | Human | PMID:21402783 |
| Molecular Function | GO:0005515 | protein binding | NPHS1 | IPI | UniProtKB:P16333 | Human | PMID:16525419 |
| Molecular Function | GO:0005515 | protein binding | NPHS1 | IPI | UniProtKB:B1Q236 | Human | PMID:21858180 |
| Molecular Function | GO:0005515 | protein binding | NPHS1 | IPI | UniProtKB:P46940 | Human | PMID:22662192 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | PR:Q7TSU7 | Mouse | PMID:21306299 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | PR:Q91X05 | Mouse | PMID:11733557 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | PR:Q8BR86 | Mouse | PMID:18752272 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | PR:Q61143 | Mouse | PMID:15924139 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | UniProtKB:B5DFH1 | Rat | PMID:15994232|RGD:8553727 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | UniProtKB:O88382 | Rat | PMID:15994232|RGD:8553727 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | UniProtKB:P16086 | Rat | PMID:15994232|RGD:8553727 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | UniProtKB:Q62915 | Rat | PMID:15994232|RGD:8553727 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | UniProtKB:Q9WUX0 | Rat | PMID:15994232|RGD:8553727 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | UniProtKB:O88382 | Rat | PMID:20534871|RGD:8553386 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | UniProtKB:Q91ZD4 | Rat | PMID:20534871|RGD:8553386 |
| Molecular Function | GO:0005515 | protein binding | Nphs1 | IPI | UniProtKB:P97710 | Rat | PMID:22747997|RGD:8553639 |
| Molecular Function | GO:0005515 | protein binding | nphs1 | IPI | ZFIN:ZDB-GENE-051102-2 | Zfish | PMID:24337247 |
| Molecular Function | GO:0019904 | protein domain specific binding | Nphs1 | IPI | RGD:1311884 | Rat | PMID:15331416|RGD:1600864 |
| Molecular Function | GO:0019904 | protein domain specific binding | Nphs1 | IPI | RGD:62004 | Rat | PMID:15331416|RGD:1600864 |
| Molecular Function | GO:0019904 | protein domain specific binding | Nphs1 | IPI | RGD:621855 | Rat | PMID:15331416|RGD:1600864 |
| Molecular Function | GO:0030507 | spectrin binding | Nphs1 | IPI | RGD:621714 | Rat | PMID:15331416|RGD:1600864 |
| Molecular Function | GO:0030507 | spectrin binding | Nphs1 | IPI | RGD:727922 | Rat | PMID:15331416|RGD:1600864 |
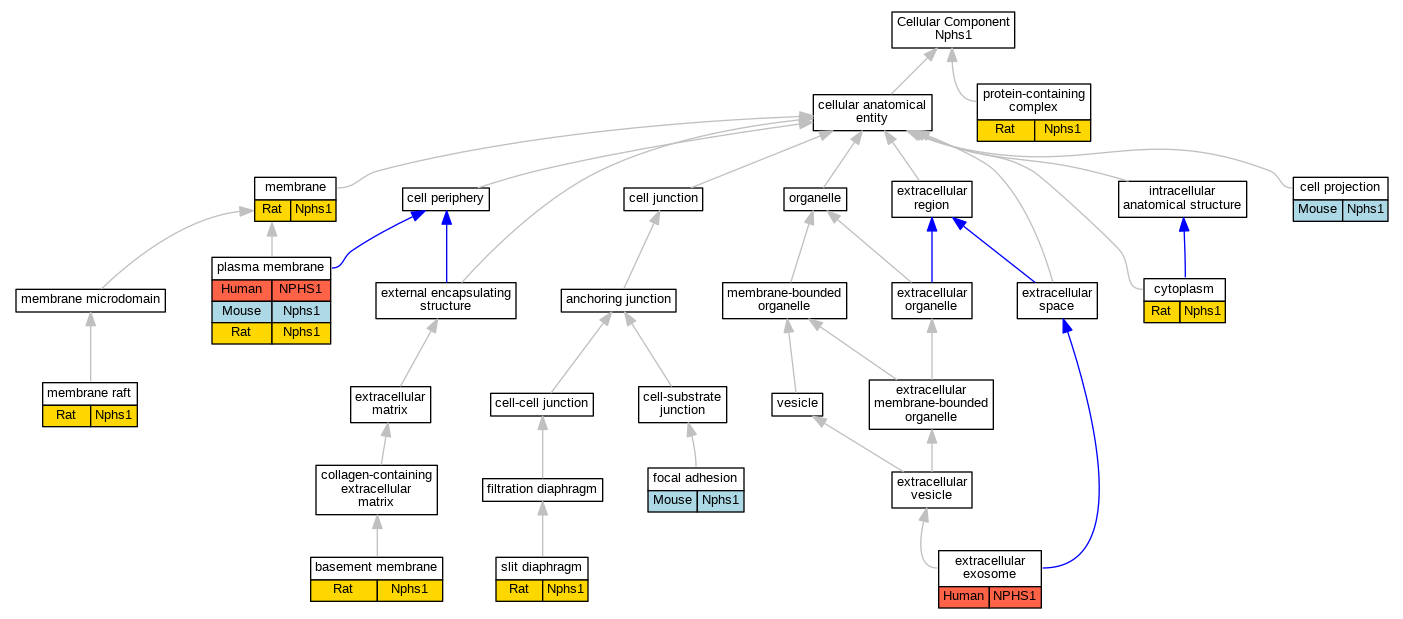
| Cellular Component | GO:0005604 | basement membrane | Nphs1 | IDA | | Rat | PMID:21876538|RGD:38599163 |
| Cellular Component | GO:0042995 | cell projection | Nphs1 | IDA | | Mouse | MGI:MGI:4887519|PMID:20534871 |
| Cellular Component | GO:0005737 | cytoplasm | Nphs1 | IDA | | Rat | PMID:21876538|RGD:38599163 |
| Cellular Component | GO:0070062 | extracellular exosome | NPHS1 | HDA | | Human | PMID:19056867 |
| Cellular Component | GO:0005925 | focal adhesion | Nphs1 | IDA | | Mouse | PMID:29162887 |
| Cellular Component | GO:0016020 | membrane | Nphs1 | IDA | | Rat | PMID:11880318|RGD:633458 |
| Cellular Component | GO:0045121 | membrane raft | Nphs1 | IDA | | Rat | PMID:11880318|RGD:633458 |
| Cellular Component | GO:0005886 | plasma membrane | NPHS1 | IDA | | Human | PMID:17464107 |
| Cellular Component | GO:0005886 | plasma membrane | Nphs1 | IDA | | Mouse | PMID:15924139 |
| Cellular Component | GO:0005886 | plasma membrane | Nphs1 | IDA | | Rat | PMID:20534871|RGD:8553386 |
| Cellular Component | GO:0032991 | protein-containing complex | Nphs1 | IDA | | Rat | PMID:15331416|RGD:1600864 |
| Cellular Component | GO:0036057 | slit diaphragm | Nphs1 | IDA | | Rat | PMID:15994232|RGD:8553727 |
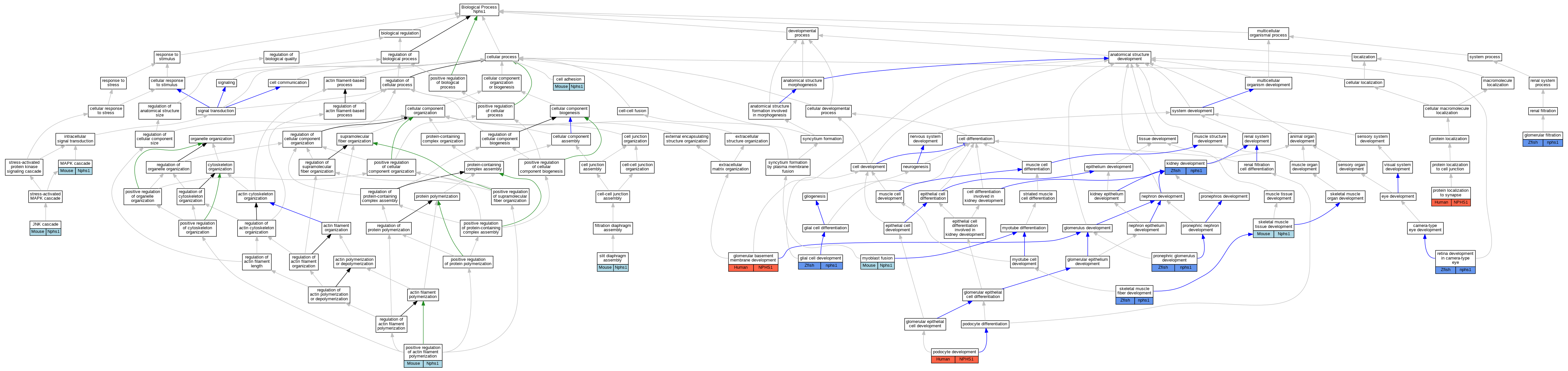
| Biological Process | GO:0007155 | cell adhesion | Nphs1 | IGI | MGI:MGI:2442334 | Mouse | PMID:21306299 |
| Biological Process | GO:0007155 | cell adhesion | Nphs1 | IGI | MGI:MGI:1891396 | Mouse | PMID:21306299 |
| Biological Process | GO:0021782 | glial cell development | nphs1 | IMP | ZFIN:ZDB-CRISPR-191009-85,ZFIN:ZDB-CRISPR-191009-86 | Zfish | PMID:30924555 |
| Biological Process | GO:0032836 | glomerular basement membrane development | NPHS1 | IEP | | Human | PMID:17464107 |
| Biological Process | GO:0003094 | glomerular filtration | nphs1 | IMP | ZFIN:ZDB-MRPHLNO-051102-1 | Zfish | PMID:25962121 |
| Biological Process | GO:0003094 | glomerular filtration | nphs1 | IMP | ZFIN:ZDB-MRPHLNO-051102-1 | Zfish | PMID:21949092 |
| Biological Process | GO:0007254 | JNK cascade | Nphs1 | IDA | | Mouse | PMID:11562357 |
| Biological Process | GO:0001822 | kidney development | nphs1 | IMP | ZFIN:ZDB-MRPHLNO-051102-1 | Zfish | PMID:16102746 |
| Biological Process | GO:0000165 | MAPK cascade | Nphs1 | IDA | | Mouse | PMID:11562357 |
| Biological Process | GO:0007520 | myoblast fusion | Nphs1 | IMP | | Mouse | MGI:MGI:3849657|PMID:19470472 |
| Biological Process | GO:0072015 | podocyte development | NPHS1 | IEP | | Human | PMID:17464107 |
| Biological Process | GO:0030838 | positive regulation of actin filament polymerization | Nphs1 | IGI | MGI:MGI:1891396 | Mouse | PMID:17923684 |
| Biological Process | GO:0030838 | positive regulation of actin filament polymerization | Nphs1 | IGI | MGI:MGI:109601 | Mouse | PMID:17923684 |
| Biological Process | GO:0030838 | positive regulation of actin filament polymerization | Nphs1 | IGI | MGI:MGI:95805 | Mouse | PMID:17923684 |
| Biological Process | GO:0039021 | pronephric glomerulus development | nphs1 | IMP | ZFIN:ZDB-MRPHLNO-140522-5 | Zfish | PMID:24337247 |
| Biological Process | GO:0039021 | pronephric glomerulus development | nphs1 | IGI | RefSeq:NM_004646.3,ZFIN:ZDB-MRPHLNO-140522-5 | Zfish | PMID:24337247 |
| Biological Process | GO:0039021 | pronephric glomerulus development | nphs1 | IMP | ZFIN:ZDB-MRPHLNO-200130-1 | Zfish | PMID:31152064 |
| Biological Process | GO:0035418 | protein localization to synapse | NPHS1 | IGI | UniProtKB:Q9U3P2 | Human | PMID:21858180 |
| Biological Process | GO:0060041 | retina development in camera-type eye | nphs1 | IMP | ZFIN:ZDB-CRISPR-191009-85,ZFIN:ZDB-CRISPR-191009-86 | Zfish | PMID:30924555 |
| Biological Process | GO:0048741 | skeletal muscle fiber development | nphs1 | IMP | ZFIN:ZDB-MRPHLNO-051102-1 | Zfish | PMID:19470472 |
| Biological Process | GO:0007519 | skeletal muscle tissue development | Nphs1 | IMP | | Mouse | MGI:MGI:3849657|PMID:19470472 |
| Biological Process | GO:0036060 | slit diaphragm assembly | Nphs1 | IMP | | Mouse | MGI:MGI:1929558|PMID:11136707 |
 Analysis Tools
Analysis Tools