|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
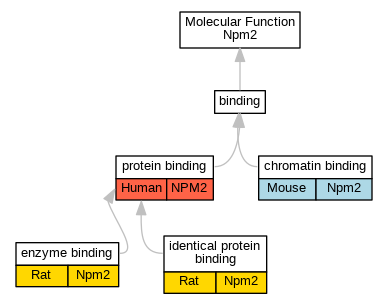
| Molecular Function | GO:0003682 | chromatin binding | Npm2 | IDA | Mouse | PMID:24105743 | |
| Molecular Function | GO:0019899 | enzyme binding | Npm2 | IDA | Rat | PMID:11481036|RGD:1600909 | |
| Molecular Function | GO:0042802 | identical protein binding | Npm2 | IPI | RGD:727816 | Rat | PMID:8611572|RGD:1600911 |
| Molecular Function | GO:0005515 | protein binding | NPM2 | IPI | UniProtKB:O75607 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | NPM2 | IPI | UniProtKB:P06748 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | NPM2 | IPI | UniProtKB:Q9H190 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | NPM2 | IPI | UniProtKB:Q9H8W4 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | NPM2 | IPI | UniProtKB:O75607 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | NPM2 | IPI | UniProtKB:O75607 | Human | PMID:31515488 |
| Molecular Function | GO:0005515 | protein binding | NPM2 | IPI | UniProtKB:P06748 | Human | PMID:31515488 |
| Molecular Function | GO:0005515 | protein binding | NPM2 | IPI | UniProtKB:Q9H190 | Human | PMID:31515488 |
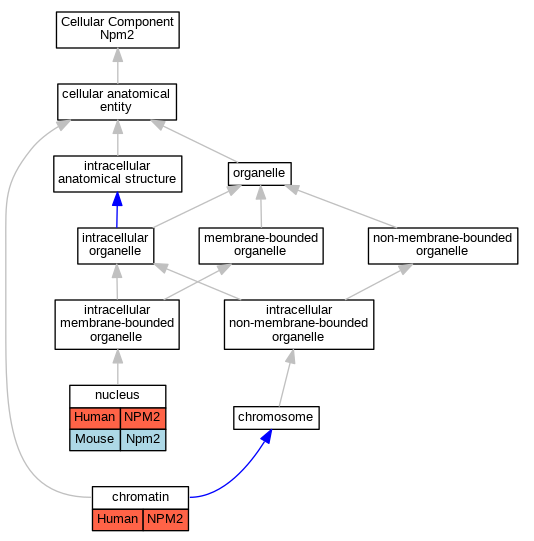
| Cellular Component | GO:0000785 | chromatin | NPM2 | IDA | Human | PMID:12714744 | |
| Cellular Component | GO:0005634 | nucleus | NPM2 | IDA | Human | PMID:12714744 | |
| Cellular Component | GO:0005634 | nucleus | Npm2 | IDA | Mouse | PMID:12714744 | |
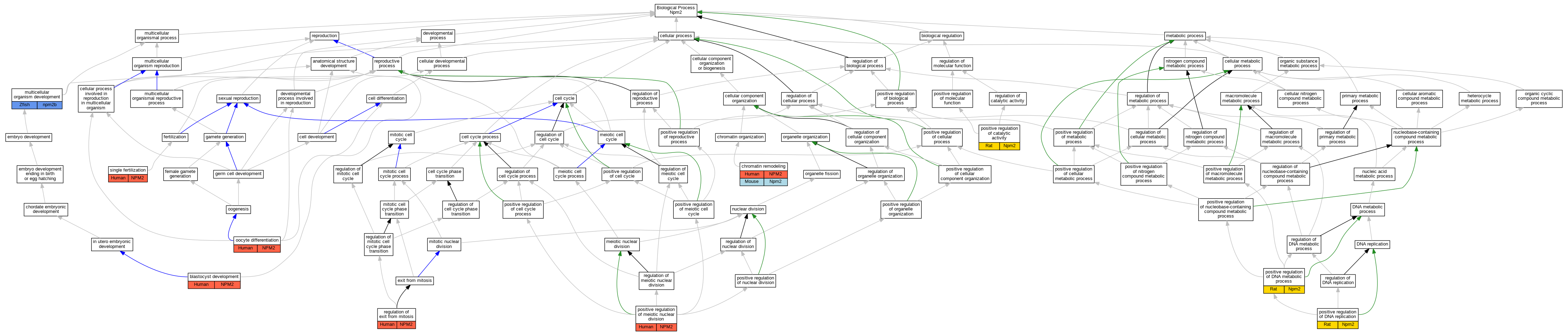
| Biological Process | GO:0001824 | blastocyst development | NPM2 | IMP | Human | PMID:12714744 | |
| Biological Process | GO:0006338 | chromatin remodeling | NPM2 | IDA | Human | PMID:12714744 | |
| Biological Process | GO:0006338 | chromatin remodeling | Npm2 | IMP | MGI:MGI:2182141 | Mouse | PMID:12714744 |
| Biological Process | GO:0007275 | multicellular organism development | npm2b | IMP | ZFIN:ZDB-MRPHLNO-151103-1 | Zfish | PMID:25009208 |
| Biological Process | GO:0009994 | oocyte differentiation | NPM2 | IEP | Human | PMID:12714744 | |
| Biological Process | GO:0043085 | positive regulation of catalytic activity | Npm2 | IDA | Rat | PMID:11481036|RGD:1600909 | |
| Biological Process | GO:0051054 | positive regulation of DNA metabolic process | Npm2 | IDA | Rat | PMID:11481036|RGD:1600909 | |
| Biological Process | GO:0045740 | positive regulation of DNA replication | Npm2 | IDA | Rat | PMID:11481036|RGD:1600909 | |
| Biological Process | GO:0045836 | positive regulation of meiotic nuclear division | NPM2 | IMP | Human | PMID:12714744 | |
| Biological Process | GO:0007096 | regulation of exit from mitosis | NPM2 | IMP | Human | PMID:12714744 | |
| Biological Process | GO:0007338 | single fertilization | NPM2 | IMP | Human | PMID:12714744 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 09/03/2024 MGI 6.24 |

|
|
|
||