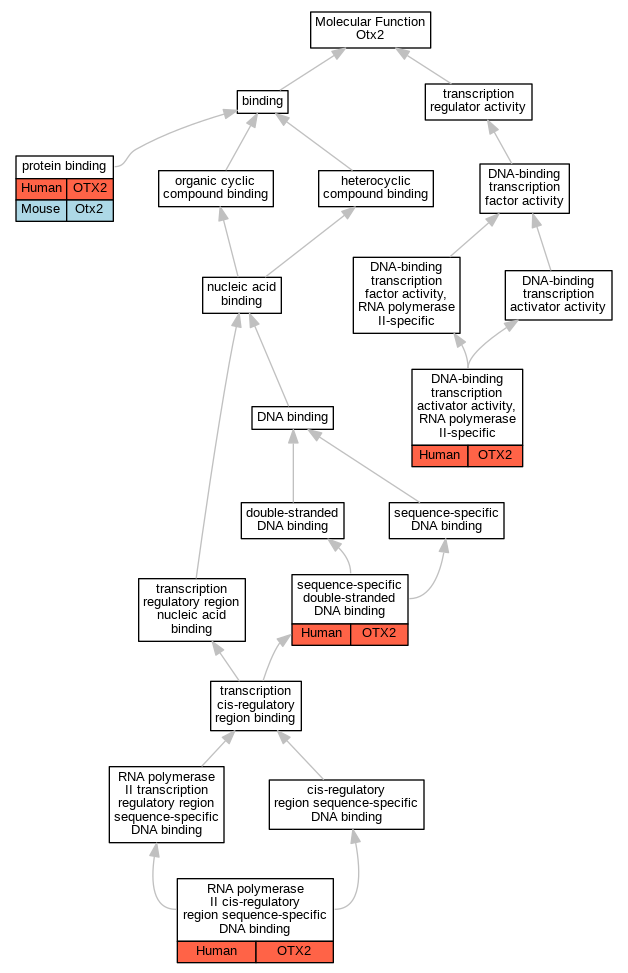
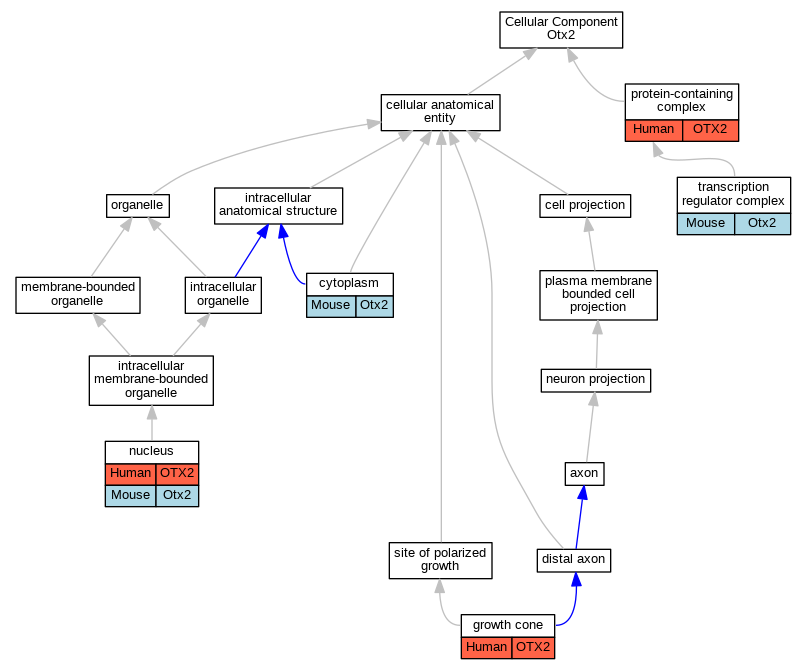
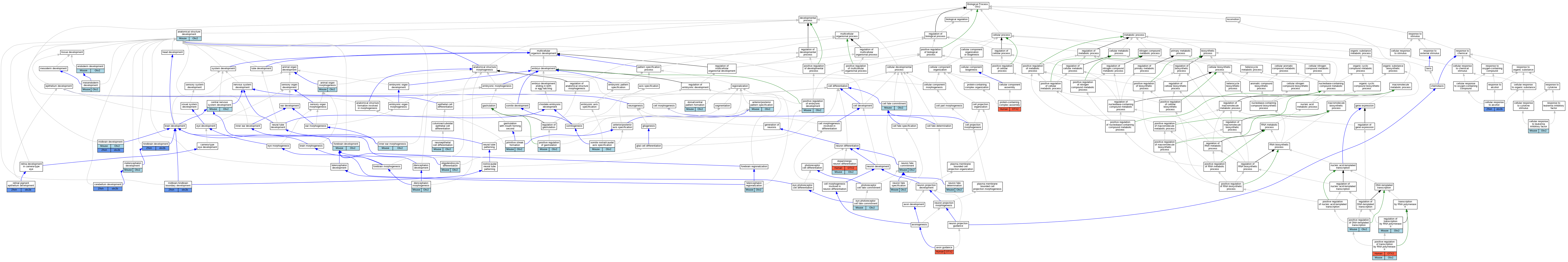
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | OTX2 | IDA | | Human | PMID:12559959 |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | OTX2 | IDA | | Human | PMID:20530484 |
| Molecular Function | GO:0005515 | protein binding | OTX2 | IPI | UniProtKB:P48436 | Human | PMID:20530484 |
| Molecular Function | GO:0005515 | protein binding | Otx2 | IPI | UniProtKB:Q62441 | Mouse | MGI:MGI:6245330|PMID:17060451 |
| Molecular Function | GO:0005515 | protein binding | Otx2 | IPI | PR:Q91ZK4 | Mouse | PMID:15890343 |
| Molecular Function | GO:0005515 | protein binding | Otx2 | IPI | PR:Q62441 | Mouse | PMID:15105370 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | OTX2 | IDA | | Human | PMID:12559959 |
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | OTX2 | IDA | | Human | PMID:28473536 |
| Cellular Component | GO:0005737 | cytoplasm | Otx2 | IDA | | Mouse | PMID:10891582 |
| Cellular Component | GO:0030426 | growth cone | OTX2 | IDA | | Human | PMID:16267555 |
| Cellular Component | GO:0005634 | nucleus | OTX2 | IDA | | Human | PMID:24399192 |
| Cellular Component | GO:0005634 | nucleus | Otx2 | IDA | | Mouse | PMID:22992956 |
| Cellular Component | GO:0005634 | nucleus | Otx2 | IDA | | Mouse | PMID:19796622 |
| Cellular Component | GO:0005634 | nucleus | Otx2 | IDA | | Mouse | PMID:10891582 |
| Cellular Component | GO:0005634 | nucleus | Otx2 | IDA | | Mouse | PMID:23056351 |
| Cellular Component | GO:0032991 | protein-containing complex | OTX2 | IDA | | Human | PMID:20530484 |
| Cellular Component | GO:0005667 | transcription regulator complex | Otx2 | IDA | | Mouse | PMID:26494787 |
| Biological Process | GO:0048856 | anatomical structure development | Otx2 | IGI | MGI:MGI:109448 | Mouse | PMID:11412024 |
| Biological Process | GO:0009887 | animal organ morphogenesis | Otx2 | IMP | MGI:MGI:2178753 | Mouse | PMID:14625556 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | Otx2 | IMP | MGI:MGI:1857502 | Mouse | PMID:9501024 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | Otx2 | IGI | MGI:MGI:109448 | Mouse | PMID:11412024 |
| Biological Process | GO:0009952 | anterior/posterior pattern specification | Otx2 | IMP | MGI:MGI:2661067 | Mouse | PMID:12652306 |
| Biological Process | GO:0007411 | axon guidance | OTX2 | IDA | | Human | PMID:16267555 |
| Biological Process | GO:0045165 | cell fate commitment | Otx2 | IMP | MGI:MGI:2661065 | Mouse | PMID:15105370 |
| Biological Process | GO:0097306 | cellular response to alcohol | otx2b | IDA | | Zfish | PMID:17013923 |
| Biological Process | GO:1990830 | cellular response to leukemia inhibitory factor | Otx2 | IEP | | Mouse | PMID:20439489 |
| Biological Process | GO:0007417 | central nervous system development | Otx2 | IMP | MGI:MGI:2150146 | Mouse | PMID:11731459 |
| Biological Process | GO:0007417 | central nervous system development | Otx2 | IMP | MGI:MGI:2661067 | Mouse | PMID:12652306 |
| Biological Process | GO:0007417 | central nervous system development | Otx2 | IMP | MGI:MGI:1857510 | Mouse | PMID:11731459 |
| Biological Process | GO:0021549 | cerebellum development | otx2b | IGI | ZFIN:ZDB-GENO-140305-1,ZFIN:ZDB-MRPHLNO-060620-3,ZFIN:ZDB-MRPHLNO-060620-5 | Zfish | PMID:24183937 |
| Biological Process | GO:0048852 | diencephalon morphogenesis | Otx2 | IGI | MGI:MGI:97450 | Mouse | PMID:20816794 |
| Biological Process | GO:0071542 | dopaminergic neuron differentiation | OTX2 | IGI | UniProtKB:Q9JKU8,UniProtKB:Q9Y261 | Human | PMID:19951692 |
| Biological Process | GO:0071542 | dopaminergic neuron differentiation | Otx2 | IMP | MGI:MGI:2176222,MGI:MGI:2661065 | Mouse | PMID:15888661 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | Otx2 | IMP | MGI:MGI:2661067 | Mouse | PMID:12652306 |
| Biological Process | GO:0007492 | endoderm development | Otx2 | IGI | MGI:MGI:109448 | Mouse | PMID:11412024 |
| Biological Process | GO:0042706 | eye photoreceptor cell fate commitment | Otx2 | IMP | MGI:MGI:2178753 | Mouse | PMID:14625556 |
| Biological Process | GO:0030900 | forebrain development | Otx2 | IGI | MGI:MGI:97450 | Mouse | PMID:15201223 |
| Biological Process | GO:0030900 | forebrain development | Otx2 | IGI | MGI:MGI:95388 | Mouse | PMID:15917450 |
| Biological Process | GO:0030900 | forebrain development | Otx2 | IMP | MGI:MGI:2178753 | Mouse | PMID:14625556 |
| Biological Process | GO:0030900 | forebrain development | Otx2 | IMP | MGI:MGI:1857510 | Mouse | PMID:15201223 |
| Biological Process | GO:0030900 | forebrain development | Otx2 | IMP | MGI:MGI:3046841 | Mouse | PMID:15201223 |
| Biological Process | GO:0030902 | hindbrain development | otx2b | IGI | ZFIN:ZDB-GENO-980202-822,ZFIN:ZDB-MRPHLNO-060620-3,ZFIN:ZDB-MRPHLNO-060620-5 | Zfish | PMID:16611693 |
| Biological Process | GO:0042472 | inner ear morphogenesis | Otx2 | IGI | MGI:MGI:97450 | Mouse | PMID:10225993 |
| Biological Process | GO:0048382 | mesendoderm development | Otx2 | IMP | MGI:MGI:5698524,MGI:MGI:2656539 | Mouse | PMID:26494787 |
| Biological Process | GO:0022037 | metencephalon development | Otx2 | IGI | MGI:MGI:97450 | Mouse | PMID:20816794 |
| Biological Process | GO:0030901 | midbrain development | Otx2 | IGI | MGI:MGI:97450 | Mouse | PMID:15201224 |
| Biological Process | GO:0030901 | midbrain development | Otx2 | IMP | MGI:MGI:3046841 | Mouse | PMID:15201223 |
| Biological Process | GO:0030901 | midbrain development | Otx2 | IMP | MGI:MGI:2176222,MGI:MGI:2661065 | Mouse | PMID:15888661 |
| Biological Process | GO:0030901 | midbrain development | Otx2 | IMP | MGI:MGI:1857510 | Mouse | PMID:15201223 |
| Biological Process | GO:0030901 | midbrain development | Otx2 | IGI | MGI:MGI:97450 | Mouse | PMID:20816794 |
| Biological Process | GO:0030901 | midbrain development | otx2b | IGI | ZFIN:ZDB-MRPHLNO-060620-3,ZFIN:ZDB-MRPHLNO-060620-5 | Zfish | PMID:16611693 |
| Biological Process | GO:0030917 | midbrain-hindbrain boundary development | otx2b | IGI | ZFIN:ZDB-MRPHLNO-060620-3,ZFIN:ZDB-MRPHLNO-060620-5 | Zfish | PMID:16611693 |
| Biological Process | GO:0060563 | neuroepithelial cell differentiation | Otx2 | IMP | MGI:MGI:2178753 | Mouse | PMID:14625556 |
| Biological Process | GO:0048663 | neuron fate commitment | Otx2 | IMP | MGI:MGI:2178753,MGI:MGI:3665330 | Mouse | PMID:26166575 |
| Biological Process | GO:0048663 | neuron fate commitment | Otx2 | IMP | MGI:MGI:2446434,MGI:MGI:2661065 | Mouse | PMID:16339193 |
| Biological Process | GO:0048664 | neuron fate determination | Otx2 | IMP | MGI:MGI:2446434,MGI:MGI:2661065 | Mouse | PMID:19592574 |
| Biological Process | GO:0048665 | neuron fate specification | Otx2 | IMP | MGI:MGI:2176222,MGI:MGI:2661065 | Mouse | PMID:15888661 |
| Biological Process | GO:0048709 | oligodendrocyte differentiation | Otx2 | IMP | MGI:MGI:2178753,MGI:MGI:3665330 | Mouse | PMID:26166575 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | Otx2 | IMP | | Mouse | MGI:MGI:2157654|PMID:11291865 |
| Biological Process | GO:0040019 | positive regulation of embryonic development | Otx2 | IMP | | Mouse | MGI:MGI:2157654|PMID:11291865 |
| Biological Process | GO:2000543 | positive regulation of gastrulation | Otx2 | IMP | | Mouse | MGI:MGI:2157654|PMID:11291865 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | OTX2 | IDA | | Human | PMID:12559959 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | OTX2 | IDA | | Human | PMID:20530484 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Otx2 | IGI | MGI:MGI:2153518 | Mouse | PMID:15890343 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Otx2 | IDA | | Mouse | PMID:16539743 |
| Biological Process | GO:0090009 | primitive streak formation | Otx2 | IMP | MGI:MGI:1857502 | Mouse | PMID:11291865 |
| Biological Process | GO:0065003 | protein-containing complex assembly | OTX2 | IDA | | Human | PMID:20530484 |
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | Otx2 | IDA | | Mouse | PMID:15105370 |
| Biological Process | GO:0003406 | retinal pigment epithelium development | otx2b | IGI | ZFIN:ZDB-MRPHLNO-060411-1,ZFIN:ZDB-MRPHLNO-060620-3,ZFIN:ZDB-MRPHLNO-060620-5 | Zfish | PMID:23139843 |
| Biological Process | GO:0003406 | retinal pigment epithelium development | otx2b | IGI | ZFIN:ZDB-MRPHLNO-060620-3,ZFIN:ZDB-MRPHLNO-060620-5 | Zfish | PMID:23139843 |
| Biological Process | GO:0003406 | retinal pigment epithelium development | otx2b | IGI | ZFIN:ZDB-GENO-130107-2,ZFIN:ZDB-MRPHLNO-060620-3,ZFIN:ZDB-MRPHLNO-060620-5 | Zfish | PMID:23139843 |
| Biological Process | GO:0032525 | somite rostral/caudal axis specification | Otx2 | IMP | MGI:MGI:1857502 | Mouse | PMID:11291865 |
| Biological Process | GO:0021978 | telencephalon regionalization | Otx2 | IMP | MGI:MGI:2178753,MGI:MGI:3665330 | Mouse | PMID:26166575 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools