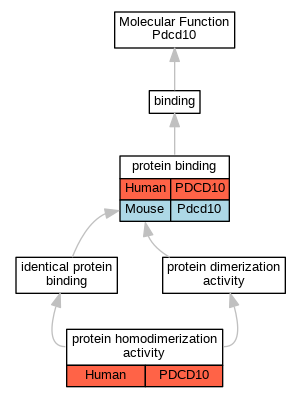
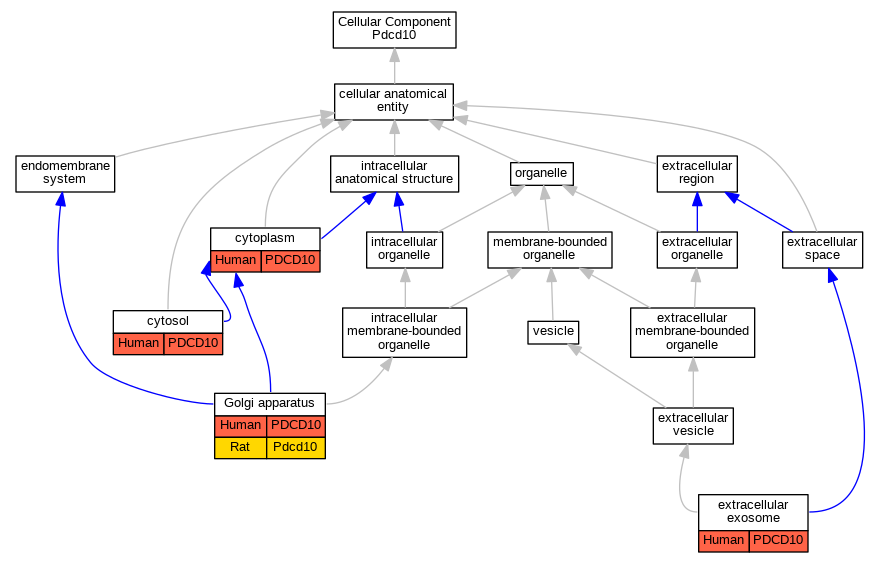
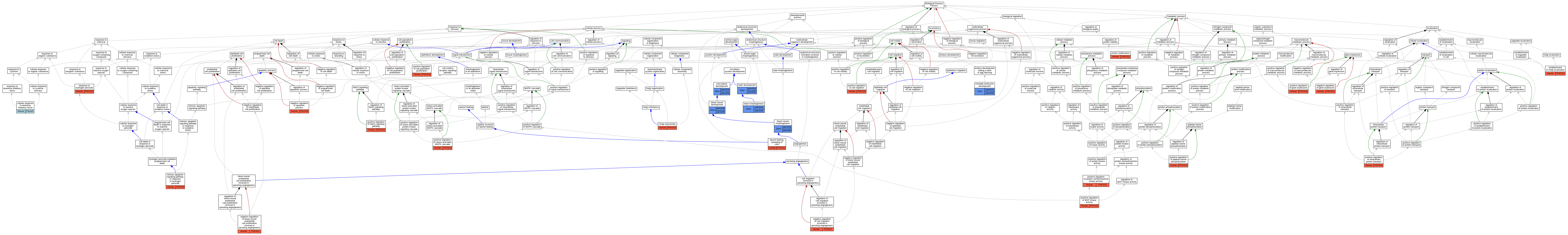
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:16189514 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0 | Human | PMID:16189514 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289 | Human | PMID:17360971 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:17657516 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q12923 | Human | PMID:17657516 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9BSQ5 | Human | PMID:17657516 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0 | Human | PMID:17657516 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:18782753 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O43815 | Human | PMID:18782753 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289 | Human | PMID:18782753 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0 | Human | PMID:18782753 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:20332113 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q08379 | Human | PMID:20332113 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289 | Human | PMID:20332113 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0 | Human | PMID:20332113 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:P49023 | Human | PMID:20489202 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9BSQ5 | Human | PMID:20489202 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:21516116 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0 | Human | PMID:21516116 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:21561863 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289 | Human | PMID:21561863 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:22652780 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9BSQ5 | Human | PMID:23266514 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:23455922 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289 | Human | PMID:23455922 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0 | Human | PMID:23455922 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289 | Human | PMID:23541896 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289-1 | Human | PMID:23541896 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0 | Human | PMID:23541896 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:23665169 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289-1 | Human | PMID:23665169 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O43815 | Human | PMID:24366813 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q499L9 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q8IY42 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q96BA2 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0-2 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O43815 | Human | PMID:26496610 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q6ZS17 | Human | PMID:27807006 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289 | Human | PMID:27807006 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0 | Human | PMID:27807006 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O43815 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q8IY42 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0-2 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:31515488 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q8IY42 | Human | PMID:31515488 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289 | Human | PMID:31515488 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0 | Human | PMID:31515488 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O43815-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O94915-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q5SXH7-1 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q86WR7-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q8IY42 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9BPY3 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9UPX6 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y285 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:O00506 | Human | PMID:32707033 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9P289 | Human | PMID:32707033 |
| Molecular Function | GO:0005515 | protein binding | PDCD10 | IPI | UniProtKB:Q9Y6E0 | Human | PMID:32707033 |
| Molecular Function | GO:0005515 | protein binding | Pdcd10 | IPI | PR:Q8K2Y9 | Mouse | PMID:22898778 |
| Molecular Function | GO:0042803 | protein homodimerization activity | PDCD10 | IPI | UniProtKB:Q9BUL8 | Human | PMID:20489202 |
| Molecular Function | GO:0042803 | protein homodimerization activity | PDCD10 | IPI | UniProtKB:Q9BUL8 | Human | PMID:21561863 |
| Cellular Component | GO:0005737 | cytoplasm | PDCD10 | IDA | | Human | PMID:22652780 |
| Cellular Component | GO:0005829 | cytosol | PDCD10 | IDA | | Human | PMID:17360971 |
| Cellular Component | GO:0005829 | cytosol | PDCD10 | IDA | | Human | PMID:20489202 |
| Cellular Component | GO:0070062 | extracellular exosome | PDCD10 | HDA | | Human | PMID:19056867 |
| Cellular Component | GO:0070062 | extracellular exosome | PDCD10 | HDA | | Human | PMID:20458337 |
| Cellular Component | GO:0070062 | extracellular exosome | PDCD10 | HDA | | Human | PMID:23533145 |
| Cellular Component | GO:0005794 | Golgi apparatus | PDCD10 | IDA | | Human | PMID:22652780 |
| Cellular Component | GO:0005794 | Golgi apparatus | PDCD10 | IDA | | Human | PMID:20332113 |
| Cellular Component | GO:0005794 | Golgi apparatus | Pdcd10 | IDA | | Rat | PMID:20332113|RGD:10047367 |
| Biological Process | GO:0001568 | blood vessel development | pdcd10a | IMP | ZFIN:ZDB-MRPHLNO-090626-2 | Zfish | PMID:19370760 |
| Biological Process | GO:0001568 | blood vessel development | pdcd10a | IGI | ZFIN:ZDB-MRPHLNO-090625-1 | Zfish | PMID:19370760 |
| Biological Process | GO:0001568 | blood vessel development | pdcd10a | IGI | ZFIN:ZDB-MRPHLNO-090626-3,ZFIN:ZDB-MRPHLNO-090626-4 | Zfish | PMID:19370760 |
| Biological Process | GO:0001568 | blood vessel development | pdcd10a | IMP | ZFIN:ZDB-MRPHLNO-090626-3 | Zfish | PMID:19370760 |
| Biological Process | GO:0001568 | blood vessel development | pdcd10a | IGI | ZFIN:ZDB-MRPHLNO-090625-1 | Zfish | PMID:22182521 |
| Biological Process | GO:0001568 | blood vessel development | pdcd10b | IGI | ZFIN:ZDB-MRPHLNO-090625-1 | Zfish | PMID:19370760 |
| Biological Process | GO:0001568 | blood vessel development | pdcd10b | IGI | ZFIN:ZDB-MRPHLNO-090626-3,ZFIN:ZDB-MRPHLNO-090626-4 | Zfish | PMID:19370760 |
| Biological Process | GO:0001568 | blood vessel development | pdcd10b | IMP | ZFIN:ZDB-MRPHLNO-090626-4 | Zfish | PMID:19370760 |
| Biological Process | GO:0001568 | blood vessel development | pdcd10b | IGI | ZFIN:ZDB-MRPHLNO-090625-1 | Zfish | PMID:22182521 |
| Biological Process | GO:0048514 | blood vessel morphogenesis | pdcd10a | IGI | ZFIN:ZDB-MRPHLNO-090625-1,ZFIN:ZDB-MRPHLNO-120220-2 | Zfish | PMID:22182521 |
| Biological Process | GO:0048514 | blood vessel morphogenesis | pdcd10b | IGI | ZFIN:ZDB-MRPHLNO-090625-1,ZFIN:ZDB-MRPHLNO-120220-2 | Zfish | PMID:22182521 |
| Biological Process | GO:1990830 | cellular response to leukemia inhibitory factor | Pdcd10 | IEP | | Mouse | PMID:20439489 |
| Biological Process | GO:0043009 | chordate embryonic development | pdcd10a | IGI | ZFIN:ZDB-MRPHLNO-101014-10 | Zfish | PMID:20592472 |
| Biological Process | GO:0043009 | chordate embryonic development | pdcd10b | IMP | ZFIN:ZDB-MRPHLNO-101014-14 | Zfish | PMID:20592472 |
| Biological Process | GO:0043009 | chordate embryonic development | pdcd10b | IGI | ZFIN:ZDB-MRPHLNO-101014-10 | Zfish | PMID:20592472 |
| Biological Process | GO:0051683 | establishment of Golgi localization | PDCD10 | IMP | | Human | PMID:20332113 |
| Biological Process | GO:0090168 | Golgi reassembly | PDCD10 | IMP | | Human | PMID:20332113 |
| Biological Process | GO:0007507 | heart development | pdcd10a | IGI | ZFIN:ZDB-MRPHLNO-101014-13,ZFIN:ZDB-MRPHLNO-101014-15 | Zfish | PMID:20592472 |
| Biological Process | GO:0007507 | heart development | pdcd10a | IGI | ZFIN:ZDB-MRPHLNO-101014-13,ZFIN:ZDB-MRPHLNO-101014-5 | Zfish | PMID:20592472 |
| Biological Process | GO:0007507 | heart development | pdcd10b | IGI | ZFIN:ZDB-MRPHLNO-101014-13,ZFIN:ZDB-MRPHLNO-101014-15 | Zfish | PMID:20592472 |
| Biological Process | GO:0003007 | heart morphogenesis | pdcd10a | IGI | ZFIN:ZDB-MRPHLNO-090625-1 | Zfish | PMID:22182521 |
| Biological Process | GO:0003007 | heart morphogenesis | pdcd10b | IGI | ZFIN:ZDB-MRPHLNO-090625-1 | Zfish | PMID:22182521 |
| Biological Process | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide | PDCD10 | IGI | UniProtKB:O00506 | Human | PMID:22652780 |
| Biological Process | GO:0043066 | negative regulation of apoptotic process | PDCD10 | IDA | | Human | PMID:17360971 |
| Biological Process | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | PDCD10 | IMP | | Human | PMID:23388056 |
| Biological Process | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | PDCD10 | IMP | | Human | PMID:23388056 |
| Biological Process | GO:0010629 | negative regulation of gene expression | PDCD10 | IMP | | Human | PMID:23388056 |
| Biological Process | GO:0030335 | positive regulation of cell migration | PDCD10 | IMP | | Human | PMID:20332113 |
| Biological Process | GO:0030335 | positive regulation of cell migration | PDCD10 | IDA | | Human | PMID:23541896 |
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | PDCD10 | IDA | | Human | PMID:17360971 |
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | PDCD10 | IDA | | Human | PMID:23541896 |
| Biological Process | GO:0010628 | positive regulation of gene expression | PDCD10 | IMP | | Human | PMID:23388056 |
| Biological Process | GO:0090316 | positive regulation of intracellular protein transport | PDCD10 | IMP | | Human | PMID:27807006 |
| Biological Process | GO:0043406 | positive regulation of MAP kinase activity | PDCD10 | IDA | | Human | PMID:17360971 |
| Biological Process | GO:0045747 | positive regulation of Notch signaling pathway | PDCD10 | IMP | | Human | PMID:23388056 |
| Biological Process | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | PDCD10 | IMP | | Human | PMID:20332113 |
| Biological Process | GO:0071902 | positive regulation of protein serine/threonine kinase activity | PDCD10 | IMP | | Human | PMID:20332113 |
| Biological Process | GO:0032874 | positive regulation of stress-activated MAPK cascade | PDCD10 | IDA | | Human | PMID:22652780 |
| Biological Process | GO:0050821 | protein stabilization | PDCD10 | IMP | | Human | PMID:20332113 |
| Biological Process | GO:0050821 | protein stabilization | PDCD10 | IMP | | Human | PMID:22652780 |
| Biological Process | GO:0042542 | response to hydrogen peroxide | PDCD10 | IDA | | Human | PMID:22291017 |
| Biological Process | GO:0001944 | vasculature development | pdcd10a | IGI | ZFIN:ZDB-MRPHLNO-101014-13,ZFIN:ZDB-MRPHLNO-101014-15 | Zfish | PMID:20592472 |
| Biological Process | GO:0001944 | vasculature development | pdcd10b | IGI | ZFIN:ZDB-MRPHLNO-101014-13,ZFIN:ZDB-MRPHLNO-101014-15 | Zfish | PMID:20592472 |
| Biological Process | GO:0044319 | wound healing, spreading of cells | PDCD10 | IMP | | Human | PMID:20332113 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools