Comparative GO Graph (mouse, human, rat, zebrafish)
Compare experimental GO annotations for Human-Mouse-Rat-Zebrafish orthologs to Mouse Pes1
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
- This class contains
- Human genes PES1
- Mouse genes Pes1
- Rat genes Pes1
- Zfish genes pes
- Annotations are indicated with nodes colored by organism:
Human,
Mouse,
Rat,
Zfish
- Relations between GO terms are indicated by colored edges "is_a"; "part_of"; "regulates"; "positively_regulates"; "negatively_regulates"
- GO annotations: Human from GOA; Mouse from MGI; Rat from RGD; Zebrafish from ZFIN.
- Only experimental annotations are displayed: EXP, IDA, IEP, IGI, IMP, IPI, HTP, HDA, HMP, HGI, HEP. Evidence codes are listed at the bottom of the page.
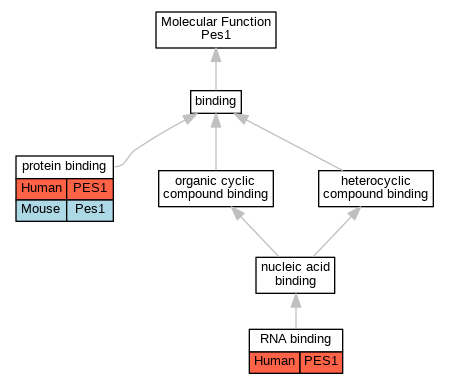
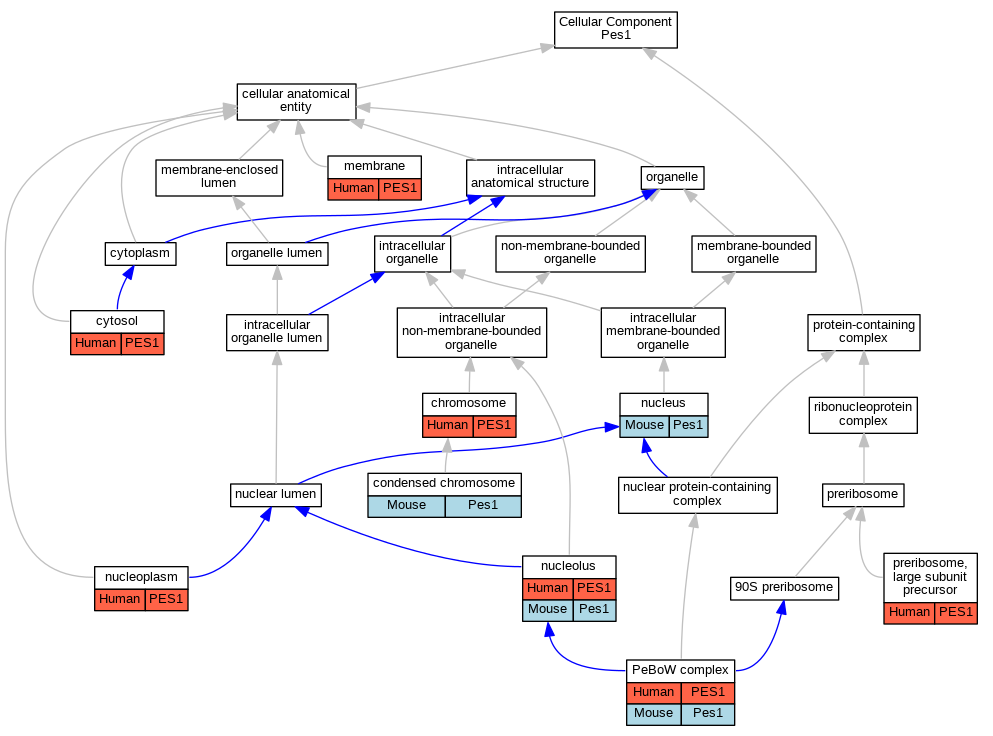
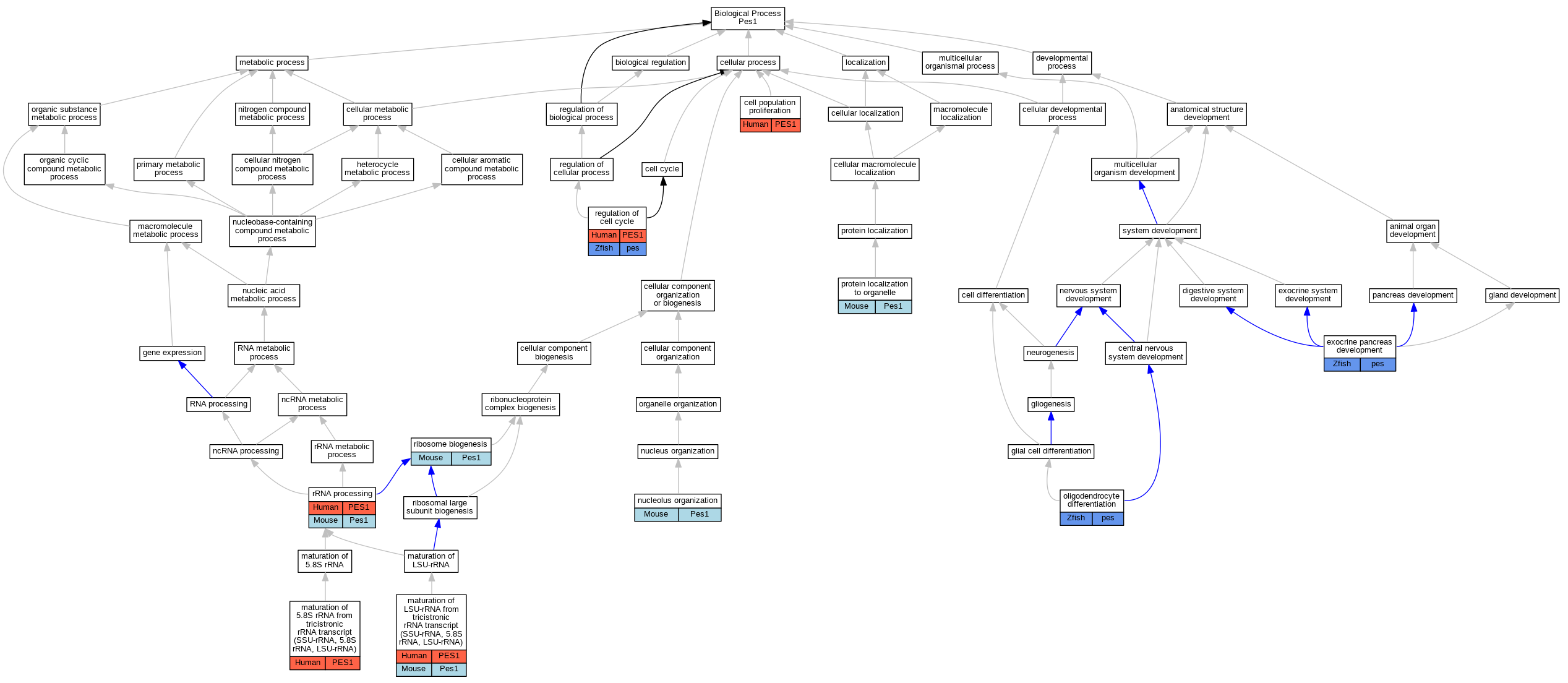 Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Pes1
Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Pes1
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:Q14137 | Human | PMID:16043514 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:Q9GZL7 | Human | PMID:16043514 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:Q14137 | Human | PMID:16738141 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:Q9GZL7 | Human | PMID:16738141 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:P97452 | Human | PMID:17353269 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:Q14137 | Human | PMID:17353269 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:Q9GZL7 | Human | PMID:17353269 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:Q08050 | Human | PMID:25609649 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:Q96GQ7 | Human | PMID:25825154 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:Q14137 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:Q9GZL7 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:Q9GZL7 | Human | PMID:30021884 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:P42858 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | PES1 | IPI | UniProtKB:P54253 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | Pes1 | IPI | PR:P14873 | Mouse | PMID:17308336 |
| Molecular Function | GO:0005515 | protein binding | Pes1 | IPI | PR:P97452 | Mouse | PMID:15225545 |
| Molecular Function | GO:0003723 | RNA binding | PES1 | HDA | | Human | PMID:22658674 |
| Molecular Function | GO:0003723 | RNA binding | PES1 | HDA | | Human | PMID:22681889 |
| Cellular Component | GO:0005694 | chromosome | PES1 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0000793 | condensed chromosome | Pes1 | IDA | | Mouse | PMID:12237316 |
| Cellular Component | GO:0005829 | cytosol | PES1 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0016020 | membrane | PES1 | HDA | | Human | PMID:19946888 |
| Cellular Component | GO:0005730 | nucleolus | PES1 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005730 | nucleolus | PES1 | IDA | | Human | PMID:16738141 |
| Cellular Component | GO:0005730 | nucleolus | Pes1 | IDA | | Mouse | PMID:12237316 |
| Cellular Component | GO:0005730 | nucleolus | Pes1 | IDA | | Mouse | PMID:15225545 |
| Cellular Component | GO:0005654 | nucleoplasm | PES1 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005654 | nucleoplasm | PES1 | IDA | | Human | PMID:16738141 |
| Cellular Component | GO:0005634 | nucleus | Pes1 | IDA | | Mouse | PMID:11112348 |
| Cellular Component | GO:0070545 | PeBoW complex | PES1 | IDA | | Human | PMID:16043514 |
| Cellular Component | GO:0070545 | PeBoW complex | PES1 | IDA | | Human | PMID:16738141 |
| Cellular Component | GO:0070545 | PeBoW complex | PES1 | IDA | | Human | PMID:17353269 |
| Cellular Component | GO:0070545 | PeBoW complex | Pes1 | IDA | | Mouse | PMID:16738141 |
| Cellular Component | GO:0030687 | preribosome, large subunit precursor | PES1 | IDA | | Human | PMID:16738141 |
| Cellular Component | GO:0030687 | preribosome, large subunit precursor | PES1 | IDA | | Human | PMID:17353269 |
| Biological Process | GO:0008283 | cell population proliferation | PES1 | IMP | | Human | PMID:16738141 |
| Biological Process | GO:0031017 | exocrine pancreas development | pes | IMP | ZFIN:ZDB-MRPHLNO-120817-3 | Zfish | PMID:22872088 |
| Biological Process | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | PES1 | IMP | | Human | PMID:16738141 |
| Biological Process | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | PES1 | IMP | | Human | PMID:16738141 |
| Biological Process | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | PES1 | IMP | | Human | PMID:17353269 |
| Biological Process | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | Pes1 | IMP | | Mouse | PMID:25190460 |
| Biological Process | GO:0007000 | nucleolus organization | Pes1 | IMP | MGI:MGI:2445464 | Mouse | PMID:12237316 |
| Biological Process | GO:0048709 | oligodendrocyte differentiation | pes | IMP | ZFIN:ZDB-GENO-120330-4 | Zfish | PMID:22384214 |
| Biological Process | GO:0033365 | protein localization to organelle | Pes1 | IMP | | Mouse | PMID:15225545 |
| Biological Process | GO:0051726 | regulation of cell cycle | PES1 | IMP | | Human | PMID:16738141 |
| Biological Process | GO:0051726 | regulation of cell cycle | pes | IMP | ZFIN:ZDB-GENO-120330-4 | Zfish | PMID:22384214 |
| Biological Process | GO:0042254 | ribosome biogenesis | Pes1 | IMP | MGI:MGI:2445464 | Mouse | PMID:12237316 |
| Biological Process | GO:0006364 | rRNA processing | PES1 | IMP | | Human | PMID:16738141 |
| Biological Process | GO:0006364 | rRNA processing | Pes1 | IGI | MGI:MGI:1334460 | Mouse | PMID:15225545 |
| Biological Process | GO:0006364 | rRNA processing | Pes1 | IMP | | Mouse | PMID:15225545 |
Gene Ontology Evidence Code Abbreviations:
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools





