| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
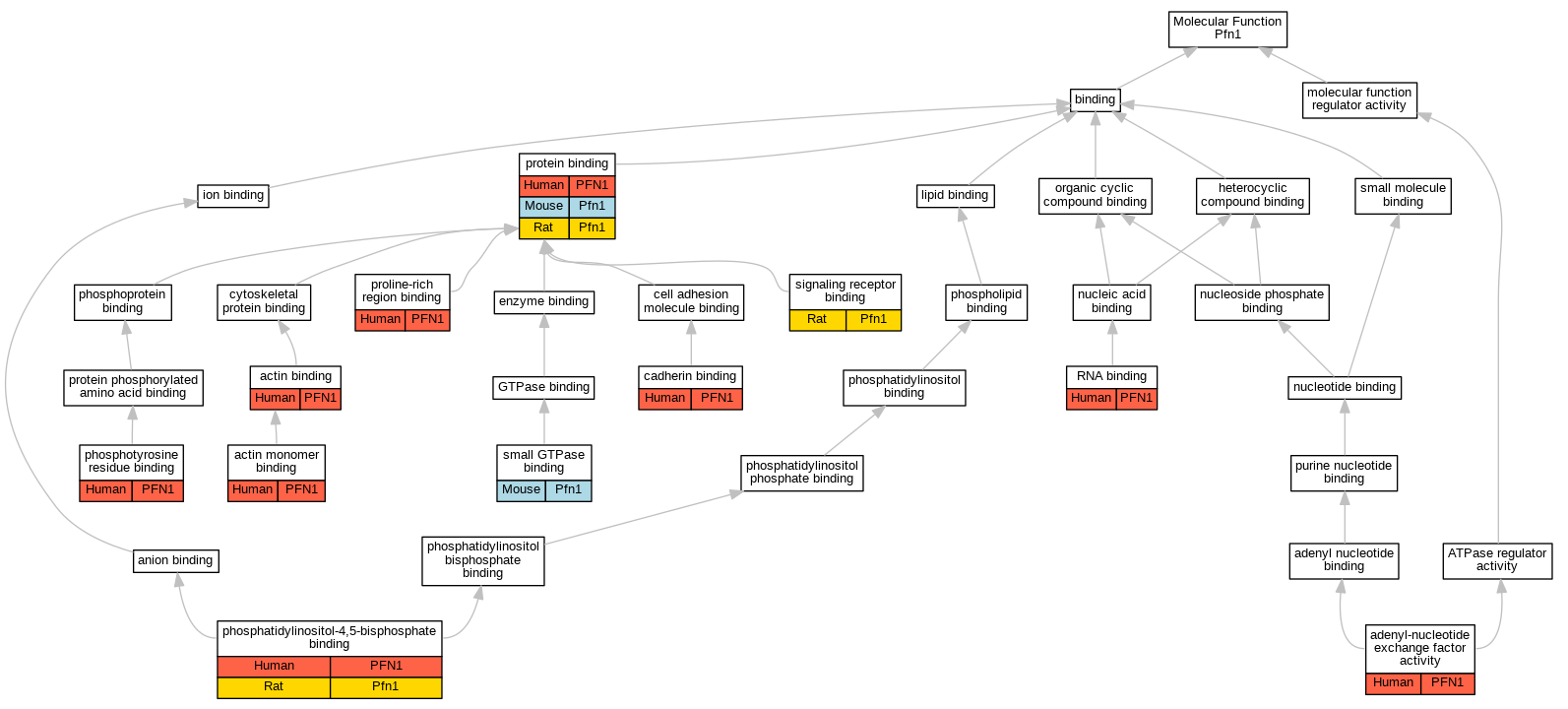
| Molecular Function | GO:0003779 | actin binding | PFN1 | IPI | UniProtKB:P68135 | Human | PMID:17914456 |
| Molecular Function | GO:0003779 | actin binding | PFN1 | IPI | UniProtKB:P68135 | Human | PMID:7758455 |
| Molecular Function | GO:0003785 | actin monomer binding | PFN1 | IDA | | Human | PMID:18573880 |
| Molecular Function | GO:0000774 | adenyl-nucleotide exchange factor activity | PFN1 | IDA | | Human | PMID:7758455 |
| Molecular Function | GO:0045296 | cadherin binding | PFN1 | HDA | | Human | PMID:25468996 |
| Molecular Function | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding | PFN1 | IDA | | Human | PMID:7758455 |
| Molecular Function | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding | Pfn1 | IDA | | Rat | PMID:16741017|RGD:2324852 |
| Molecular Function | GO:0001784 | phosphotyrosine residue binding | PFN1 | IPI | UniProtKB:Q14247 | Human | PMID:24700464 |
| Molecular Function | GO:0070064 | proline-rich region binding | PFN1 | IPI | UniProtKB:P50552 | Human | PMID:17914456 |
| Molecular Function | GO:0005515 | protein binding | PFN1 | IPI | UniProtKB:P68135 | Human | PMID:16531231 |
| Molecular Function | GO:0005515 | protein binding | PFN1 | IPI | UniProtKB:P68135 | Human | PMID:17470807 |
| Molecular Function | GO:0005515 | protein binding | PFN1 | IPI | UniProtKB:P42858 | Human | PMID:18573880 |
| Molecular Function | GO:0005515 | protein binding | PFN1 | IPI | UniProtKB:P07830 | Human | PMID:18689676 |
| Molecular Function | GO:0005515 | protein binding | PFN1 | IPI | UniProtKB:P60709 | Human | PMID:19000816 |
| Molecular Function | GO:0005515 | protein binding | PFN1 | IPI | UniProtKB:P60709 | Human | PMID:19338310 |
| Molecular Function | GO:0005515 | protein binding | PFN1 | IPI | UniProtKB:P50552 | Human | PMID:23153535 |
| Molecular Function | GO:0005515 | protein binding | PFN1 | IPI | UniProtKB:P70429 | Human | PMID:23153535 |
| Molecular Function | GO:0005515 | protein binding | PFN1 | IPI | UniProtKB:O08816 | Human | PMID:9822597 |
| Molecular Function | GO:0005515 | protein binding | PFN1 | IPI | UniProtKB:Q92558 | Human | PMID:9843499 |
| Molecular Function | GO:0005515 | protein binding | Pfn1 | IPI | UniProtKB:P03372 | Mouse | MGI:MGI:5514125|PMID:23576398 |
| Molecular Function | GO:0005515 | protein binding | Pfn1 | IPI | PR:Q8R4U1 | Mouse | PMID:15615774 |
| Molecular Function | GO:0005515 | protein binding | Pfn1 | IPI | PR:Q0QWG9 | Mouse | PMID:11826110 |
| Molecular Function | GO:0005515 | protein binding | Pfn1 | IPI | RGD:69194 | Rat | PMID:12967995|RGD:2324853 |
| Molecular Function | GO:0003723 | RNA binding | PFN1 | HDA | | Human | PMID:22681889 |
| Molecular Function | GO:0005102 | signaling receptor binding | Pfn1 | IDA | | Rat | PMID:10966486|RGD:2324858 |
| Molecular Function | GO:0031267 | small GTPase binding | Pfn1 | IPI | UniProtKB:P17081 | Mouse | MGI:MGI:3843447|PMID:10445846 |
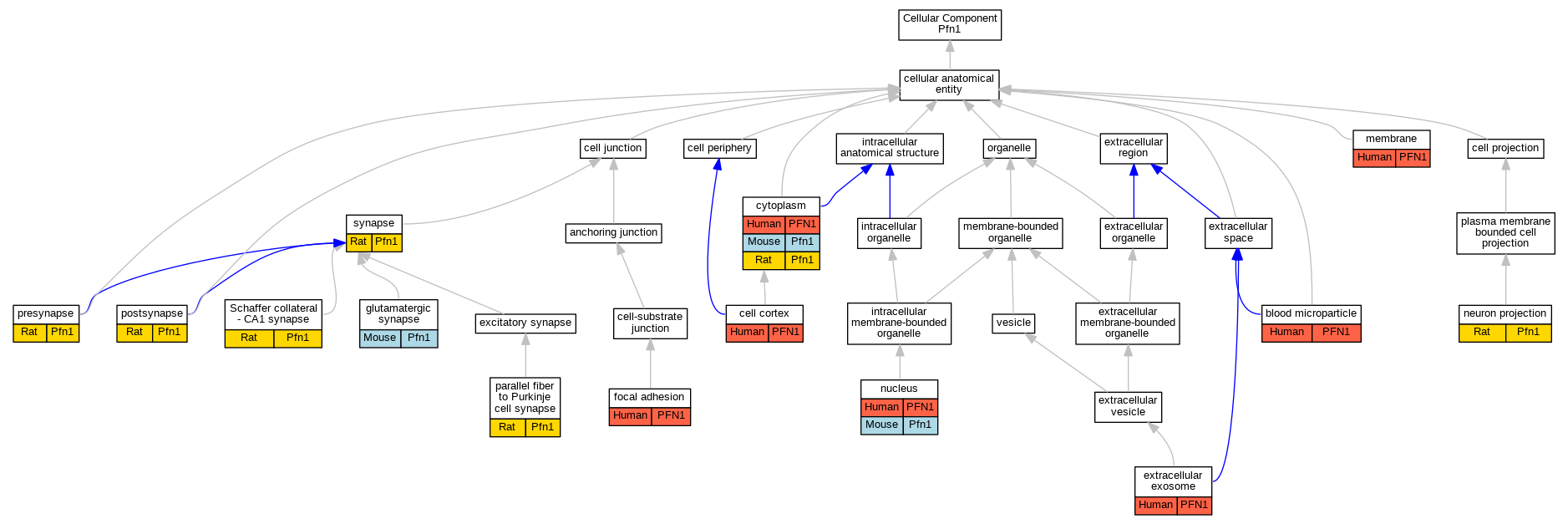
| Cellular Component | GO:0072562 | blood microparticle | PFN1 | HDA | | Human | PMID:22516433 |
| Cellular Component | GO:0005938 | cell cortex | PFN1 | IDA | | Human | PMID:24700464 |
| Cellular Component | GO:0005737 | cytoplasm | PFN1 | IDA | | Human | PMID:24700464 |
| Cellular Component | GO:0005737 | cytoplasm | Pfn1 | IDA | | Mouse | PMID:11274401 |
| Cellular Component | GO:0005737 | cytoplasm | Pfn1 | IDA | | Rat | PMID:12967995|RGD:2324853 |
| Cellular Component | GO:0070062 | extracellular exosome | PFN1 | HDA | | Human | PMID:19056867 |
| Cellular Component | GO:0070062 | extracellular exosome | PFN1 | HDA | | Human | PMID:20458337 |
| Cellular Component | GO:0070062 | extracellular exosome | PFN1 | HDA | | Human | PMID:23533145 |
| Cellular Component | GO:0005925 | focal adhesion | PFN1 | HDA | | Human | PMID:21423176 |
| Cellular Component | GO:0098978 | glutamatergic synapse | Pfn1 | IMP | | Mouse | PMID:26951674 |
| Cellular Component | GO:0098978 | glutamatergic synapse | Pfn1 | IDA | | Mouse | PMID:26951674 |
| Cellular Component | GO:0016020 | membrane | PFN1 | HDA | | Human | PMID:19946888 |
| Cellular Component | GO:0043005 | neuron projection | Pfn1 | IDA | | Rat | PMID:8106880|RGD:2324862 |
| Cellular Component | GO:0005634 | nucleus | PFN1 | IDA | | Human | PMID:24700464 |
| Cellular Component | GO:0005634 | nucleus | Pfn1 | IDA | | Mouse | PMID:15615774 |
| Cellular Component | GO:0098688 | parallel fiber to Purkinje cell synapse | Pfn1 | IDA | | Rat | PMID:15654839|RGD:13702446 |
| Cellular Component | GO:0098794 | postsynapse | Pfn1 | IDA | | Rat | PMID:15654839|RGD:13702446 |
| Cellular Component | GO:0098793 | presynapse | Pfn1 | IDA | | Rat | PMID:15654839|RGD:13702446 |
| Cellular Component | GO:0098685 | Schaffer collateral - CA1 synapse | Pfn1 | IDA | | Rat | PMID:15654839|RGD:13702446 |
| Cellular Component | GO:0045202 | synapse | Pfn1 | IDA | | Rat | PMID:12967995|RGD:2324853 |
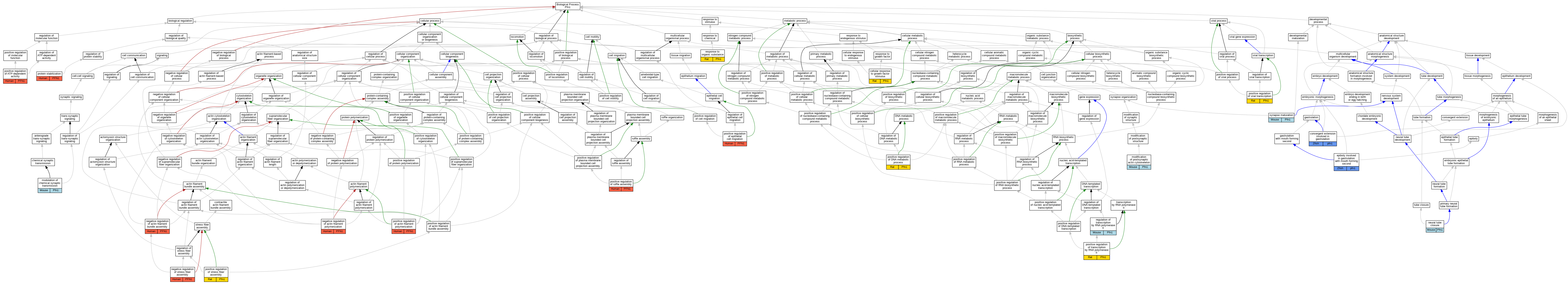
| Biological Process | GO:0071363 | cellular response to growth factor stimulus | Pfn1 | IEP | | Rat | PMID:10703666|RGD:2324859 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | pfn1 | IGI | ZFIN:ZDB-MRPHLNO-090115-2,ZFIN:ZDB-MRPHLNO-090115-3 | Zfish | PMID:18941507 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | pfn1 | IMP | ZFIN:ZDB-MRPHLNO-090115-3 | Zfish | PMID:18941507 |
| Biological Process | GO:0055113 | epiboly involved in gastrulation with mouth forming second | pfn1 | IMP | ZFIN:ZDB-MRPHLNO-090115-3 | Zfish | PMID:18941507 |
| Biological Process | GO:0055113 | epiboly involved in gastrulation with mouth forming second | pfn1 | IGI | ZFIN:ZDB-MRPHLNO-090115-2,ZFIN:ZDB-MRPHLNO-090115-3 | Zfish | PMID:18941507 |
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | Pfn1 | IDA | | Mouse | PMID:26951674 |
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | Pfn1 | IMP | | Mouse | PMID:26951674 |
| Biological Process | GO:0050804 | modulation of chemical synaptic transmission | Pfn1 | IDA | | Mouse | PMID:26951674 |
| Biological Process | GO:0050804 | modulation of chemical synaptic transmission | Pfn1 | IMP | | Mouse | PMID:26951674 |
| Biological Process | GO:0032232 | negative regulation of actin filament bundle assembly | PFN1 | IMP | | Human | PMID:23153535 |
| Biological Process | GO:0030837 | negative regulation of actin filament polymerization | PFN1 | IDA | | Human | PMID:7758455 |
| Biological Process | GO:0051497 | negative regulation of stress fiber assembly | PFN1 | IMP | | Human | PMID:23153535 |
| Biological Process | GO:0001843 | neural tube closure | Pfn1 | IGI | MGI:MGI:108360 | Mouse | PMID:10069337 |
| Biological Process | GO:0030838 | positive regulation of actin filament polymerization | PFN1 | IGI | UniProtKB:P35080 | Human | PMID:23153535 |
| Biological Process | GO:0032781 | positive regulation of ATP-dependent activity | PFN1 | IDA | | Human | PMID:7758455 |
| Biological Process | GO:0051054 | positive regulation of DNA metabolic process | Pfn1 | IDA | | Rat | PMID:10966486|RGD:2324858 |
| Biological Process | GO:0010634 | positive regulation of epithelial cell migration | PFN1 | IMP | | Human | PMID:23153535 |
| Biological Process | GO:1900029 | positive regulation of ruffle assembly | PFN1 | IMP | | Human | PMID:23153535 |
| Biological Process | GO:0051496 | positive regulation of stress fiber assembly | Pfn1 | IMP | | Rat | PMID:12740026|RGD:2324854 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Pfn1 | IDA | | Rat | PMID:10966486|RGD:2324858 |
| Biological Process | GO:0050434 | positive regulation of viral transcription | Pfn1 | IMP | | Rat | PMID:12740026|RGD:2324854 |
| Biological Process | GO:0050821 | protein stabilization | PFN1 | IDA | | Human | PMID:18573880 |
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | Pfn1 | IGI | MGI:MGI:2446472 | Mouse | PMID:15615774 |
| Biological Process | GO:0010033 | response to organic substance | Pfn1 | IEP | | Rat | PMID:8297366|RGD:2324861 |
| Biological Process | GO:0060074 | synapse maturation | Pfn1 | IDA | | Mouse | PMID:26951674 |
| Biological Process | GO:0060074 | synapse maturation | Pfn1 | IMP | | Mouse | PMID:26951674 |
 Analysis Tools
Analysis Tools