|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
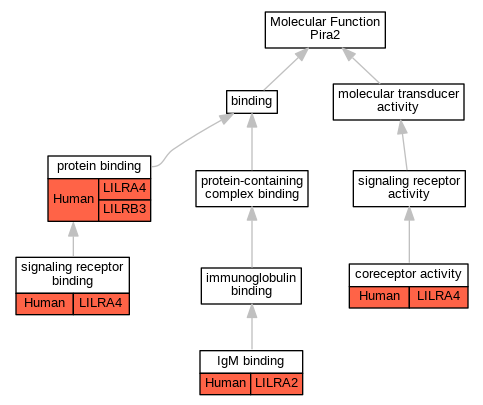
| Molecular Function | GO:0015026 | coreceptor activity | LILRA4 | IDA | Human | PMID:16735691 | |
| Molecular Function | GO:0001791 | IgM binding | LILRA2 | IDA | Human | PMID:27572839 | |
| Molecular Function | GO:0005515 | protein binding | LILRB3 | IPI | UniProtKB:P29350 | Human | PMID:18802077 |
| Molecular Function | GO:0005515 | protein binding | LILRB3 | IPI | UniProtKB:P05783 | Human | PMID:26769854 |
| Molecular Function | GO:0005515 | protein binding | LILRB3 | IPI | UniProtKB:P05787 | Human | PMID:26769854 |
| Molecular Function | GO:0005515 | protein binding | LILRA4 | IPI | UniProtKB:Q10589 | Human | PMID:19564354 |
| Molecular Function | GO:0005102 | signaling receptor binding | LILRA4 | IPI | UniProtKB:P30273 | Human | PMID:16735691 |
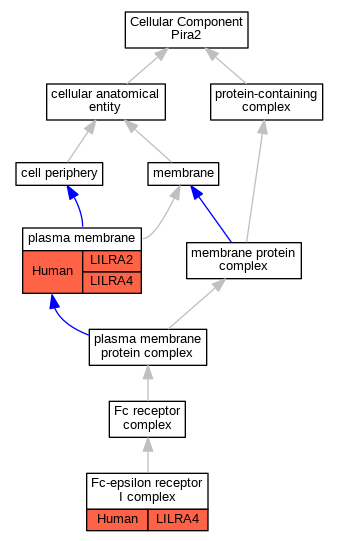
| Cellular Component | GO:0032998 | Fc-epsilon receptor I complex | LILRA4 | IDA | Human | PMID:16735691 | |
| Cellular Component | GO:0005886 | plasma membrane | LILRA4 | IDA | Human | PMID:16735691 | |
| Cellular Component | GO:0005886 | plasma membrane | LILRA2 | IDA | Human | PMID:22479404 | |
| Cellular Component | GO:0005886 | plasma membrane | LILRA2 | IDA | Human | PMID:27572839 | |
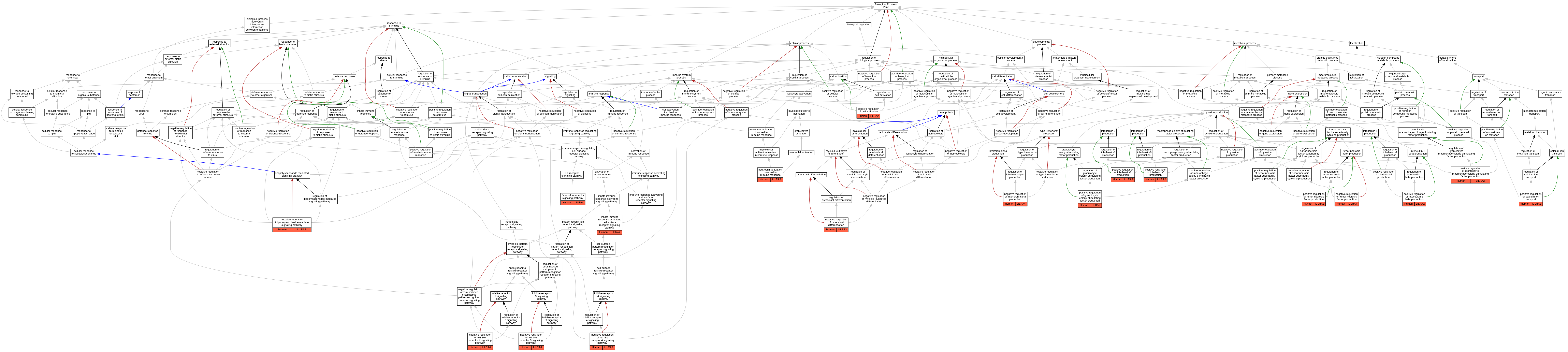
| Biological Process | GO:0038095 | Fc-epsilon receptor signaling pathway | LILRA4 | IMP | Human | PMID:16735691 | |
| Biological Process | GO:0002220 | innate immune response activating cell surface receptor signaling pathway | LILRA2 | IDA | Human | PMID:27572839 | |
| Biological Process | GO:0032687 | negative regulation of interferon-alpha production | LILRA4 | IMP | Human | PMID:16735691 | |
| Biological Process | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway | LILRA2 | IMP | Human | PMID:22479404 | |
| Biological Process | GO:0045671 | negative regulation of osteoclast differentiation | LILRB3 | IDA | Human | PMID:18802077 | |
| Biological Process | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway | LILRA2 | IMP | Human | PMID:22479404 | |
| Biological Process | GO:0034156 | negative regulation of toll-like receptor 7 signaling pathway | LILRA4 | IMP | Human | PMID:16735691 | |
| Biological Process | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway | LILRA4 | IMP | Human | PMID:16735691 | |
| Biological Process | GO:0032720 | negative regulation of tumor necrosis factor production | LILRA4 | IMP | Human | PMID:16735691 | |
| Biological Process | GO:0002283 | neutrophil activation involved in immune response | LILRA2 | IDA | Human | PMID:27572839 | |
| Biological Process | GO:0051928 | positive regulation of calcium ion transport | LILRA2 | IDA | Human | PMID:12393390 | |
| Biological Process | GO:0050867 | positive regulation of cell activation | LILRA2 | IDA | Human | PMID:12393390 | |
| Biological Process | GO:0071657 | positive regulation of granulocyte colony-stimulating factor production | LILRA2 | IMP | Human | PMID:22479404 | |
| Biological Process | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production | LILRA2 | IMP | Human | PMID:22479404 | |
| Biological Process | GO:0032731 | positive regulation of interleukin-1 beta production | LILRA2 | IDA | Human | PMID:12393390 | |
| Biological Process | GO:0032755 | positive regulation of interleukin-6 production | LILRA2 | IDA | Human | PMID:12393390 | |
| Biological Process | GO:0032755 | positive regulation of interleukin-6 production | LILRA2 | IMP | Human | PMID:22479404 | |
| Biological Process | GO:0032757 | positive regulation of interleukin-8 production | LILRA2 | IMP | Human | PMID:22479404 | |
| Biological Process | GO:0032760 | positive regulation of tumor necrosis factor production | LILRA2 | IDA | Human | PMID:12393390 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 08/02/2024 MGI 6.24 |

|
|
|
||