|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
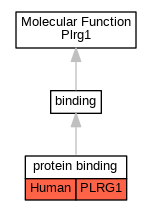
| Molecular Function | GO:0005515 | protein binding | PLRG1 | IPI | UniProtKB:O75934 | Human | PMID:20176811 |
| Molecular Function | GO:0005515 | protein binding | PLRG1 | IPI | UniProtKB:Q99459 | Human | PMID:20176811 |
| Molecular Function | GO:0005515 | protein binding | PLRG1 | IPI | UniProtKB:Q9UMS4 | Human | PMID:20176811 |
| Molecular Function | GO:0005515 | protein binding | PLRG1 | IPI | UniProtKB:P52272 | Human | PMID:20467437 |
| Molecular Function | GO:0005515 | protein binding | PLRG1 | IPI | UniProtKB:Q99459 | Human | PMID:20467437 |
| Molecular Function | GO:0005515 | protein binding | PLRG1 | IPI | UniProtKB:Q9BQ65 | Human | PMID:23022480 |
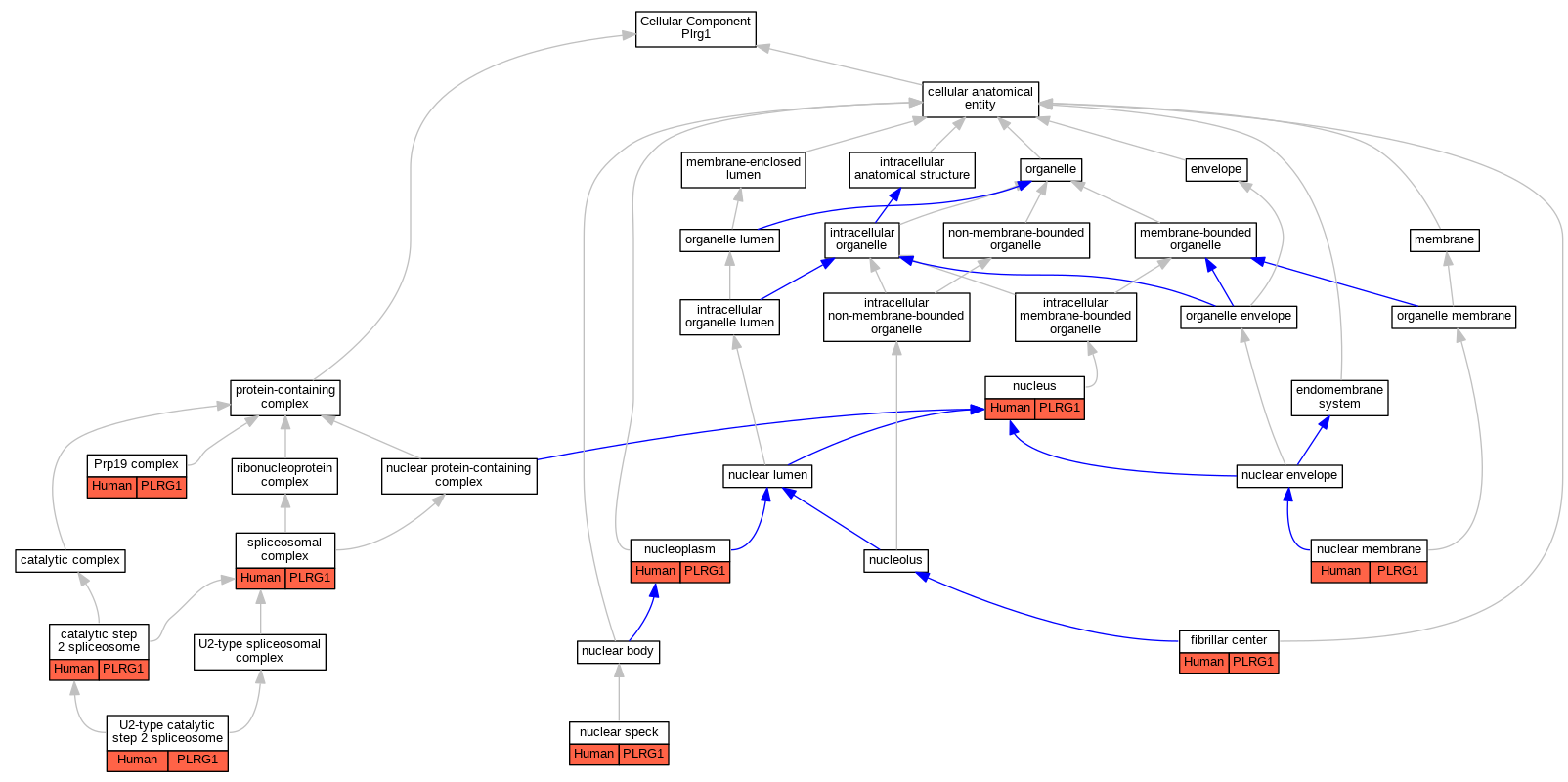
| Cellular Component | GO:0071013 | catalytic step 2 spliceosome | PLRG1 | IDA | Human | PMID:11991638 | |
| Cellular Component | GO:0001650 | fibrillar center | PLRG1 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0031965 | nuclear membrane | PLRG1 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0016607 | nuclear speck | PLRG1 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005654 | nucleoplasm | PLRG1 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005634 | nucleus | PLRG1 | IDA | Human | PMID:28076346 | |
| Cellular Component | GO:0000974 | Prp19 complex | PLRG1 | IPI | Human | PMID:20176811 | |
| Cellular Component | GO:0005681 | spliceosomal complex | PLRG1 | IPI | Human | PMID:20176811 | |
| Cellular Component | GO:0071007 | U2-type catalytic step 2 spliceosome | PLRG1 | IDA | Human | PMID:28076346 | |
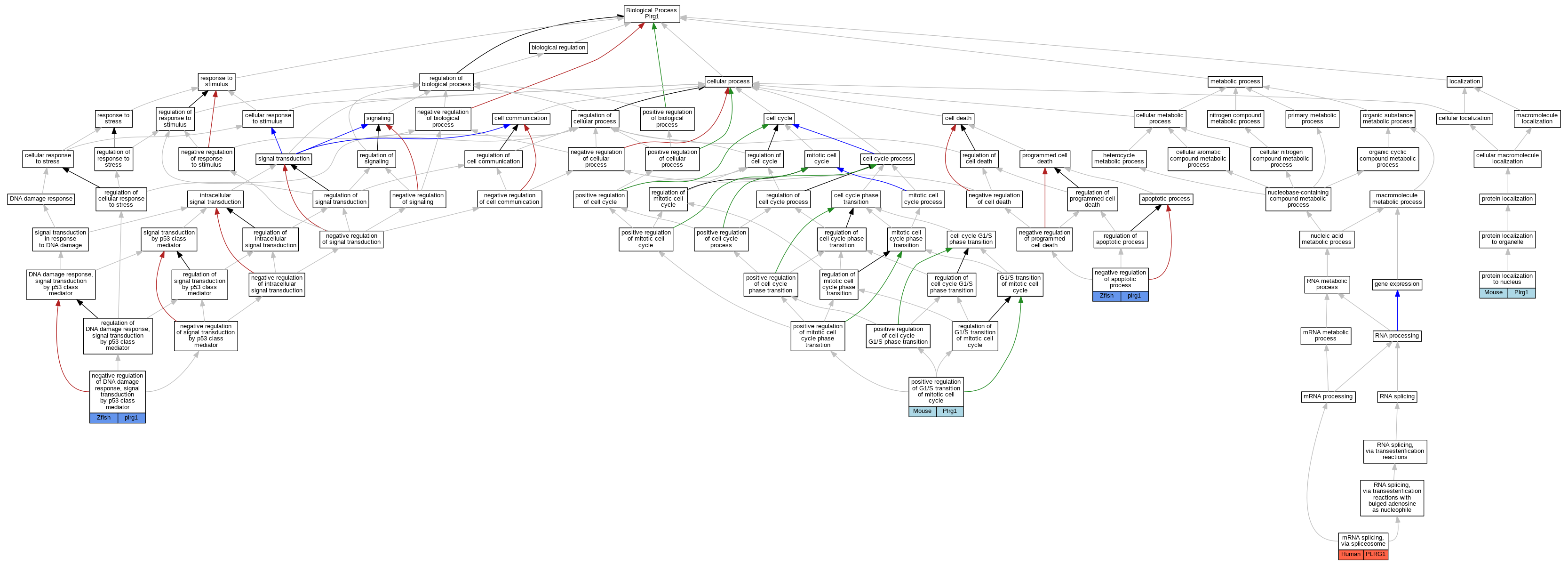
| Biological Process | GO:0000398 | mRNA splicing, via spliceosome | PLRG1 | IDA | Human | PMID:28076346 | |
| Biological Process | GO:0043066 | negative regulation of apoptotic process | plrg1 | IMP | ZFIN:ZDB-MRPHLNO-090610-4 | Zfish | PMID:19307306 |
| Biological Process | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | plrg1 | IMP | ZFIN:ZDB-MRPHLNO-090610-4 | Zfish | PMID:22952453 |
| Biological Process | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | plrg1 | IGI | ZFIN:ZDB-MRPHLNO-090610-4,ZFIN:ZDB-MRPHLNO-120919-1 | Zfish | PMID:22952453 |
| Biological Process | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | Plrg1 | IMP | MGI:MGI:3848230 | Mouse | PMID:19307306 |
| Biological Process | GO:0034504 | protein localization to nucleus | Plrg1 | IMP | MGI:MGI:3848230 | Mouse | PMID:19307306 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 06/12/2024 MGI 6.13 |

|
|
|
||