|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
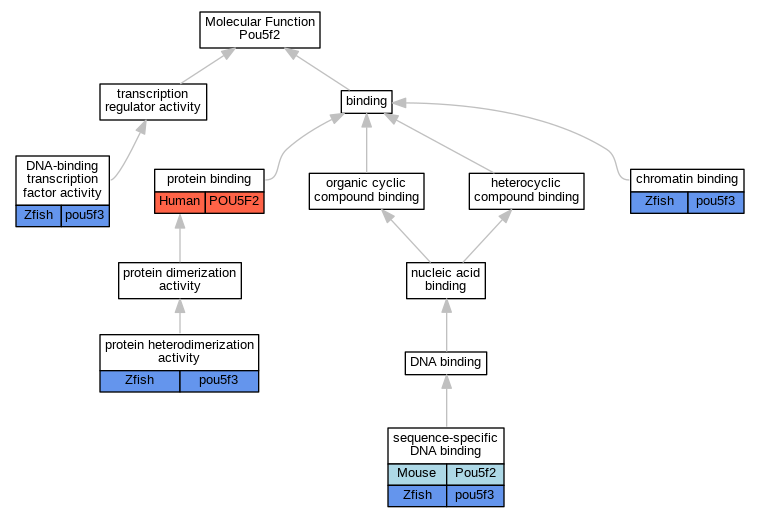
| Molecular Function | GO:0003682 | chromatin binding | pou5f3 | IDA | Zfish | PMID:30674556 | |
| Molecular Function | GO:0003682 | chromatin binding | pou5f3 | IDA | Zfish | PMID:24643012 | |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | pou5f3 | IGI | ZFIN:ZDB-GENE-011026-1 | Zfish | PMID:14723850 |
| Molecular Function | GO:0005515 | protein binding | POU5F2 | IPI | UniProtKB:Q3LI66 | Human | PMID:32296183 |
| Molecular Function | GO:0046982 | protein heterodimerization activity | pou5f3 | IPI | ZFIN:ZDB-GENE-010108-1 | Zfish | PMID:23364327 |
| Molecular Function | GO:0046982 | protein heterodimerization activity | pou5f3 | IPI | ZFIN:ZDB-GENE-011026-1 | Zfish | PMID:23364327 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | Pou5f2 | IDA | Mouse | PMID:7902581 | |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | pou5f3 | IPI | ZFIN:ZDB-GENE-980526-102 | Zfish | PMID:23950494 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | pou5f3 | IPI | ZFIN:ZDB-GENE-030909-1 | Zfish | PMID:23950494 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | pou5f3 | IDA | Zfish | PMID:23950494 | |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | pou5f3 | IPI | ZFIN:ZDB-GENE-980526-485 | Zfish | PMID:23950494 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | pou5f3 | IPI | ZFIN:ZDB-GENE-980526-333 | Zfish | PMID:23950494 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | pou5f3 | IPI | ZFIN:ZDB-GENE-980605-4 | Zfish | PMID:23950494 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | pou5f3 | IPI | ZFIN:ZDB-GENE-010111-1 | Zfish | PMID:23950494 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | pou5f3 | IPI | ZFIN:ZDB-GENE-030131-5486 | Zfish | PMID:23950494 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | pou5f3 | IPI | ZFIN:ZDB-GENE-010416-1 | Zfish | PMID:23950494 |
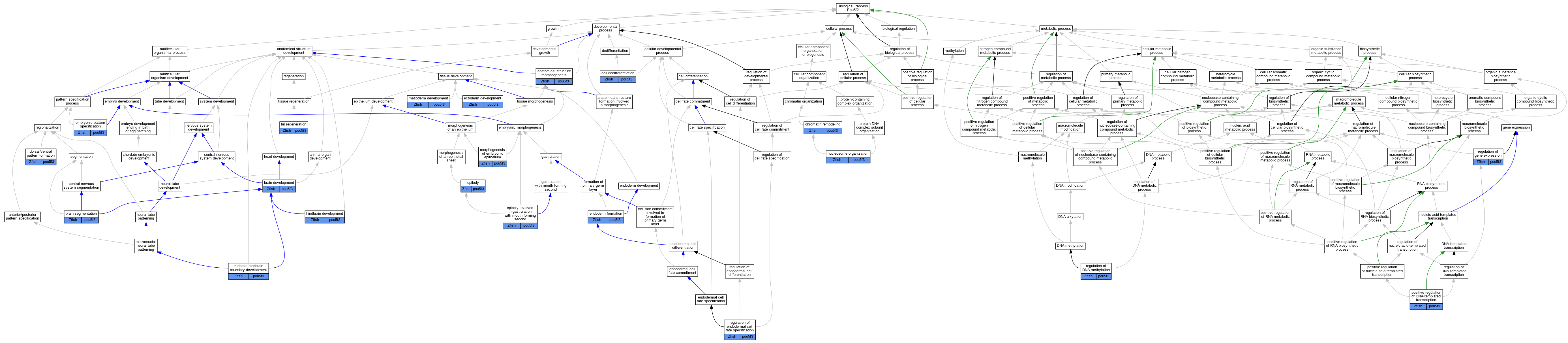
| Biological Process | GO:0009653 | anatomical structure morphogenesis | pou5f3 | IMP | ZFIN:ZDB-GENO-020625-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0009653 | anatomical structure morphogenesis | pou5f3 | IMP | ZFIN:ZDB-GENO-020426-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0007420 | brain development | pou5f3 | IMP | ZFIN:ZDB-GENO-020625-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0007420 | brain development | pou5f3 | IMP | ZFIN:ZDB-GENO-020426-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0007420 | brain development | pou5f3 | IMP | ZFIN:ZDB-GENO-050317-6 | Zfish | PMID:15593329 |
| Biological Process | GO:0035284 | brain segmentation | pou5f3 | IMP | ZFIN:ZDB-GENO-980202-270 | Zfish | PMID:11923201 |
| Biological Process | GO:0035284 | brain segmentation | pou5f3 | IMP | ZFIN:ZDB-GENO-020117-1 | Zfish | PMID:11923201 |
| Biological Process | GO:0043697 | cell dedifferentiation | pou5f3 | IMP | ZFIN:ZDB-MRPHLNO-200330-1 | Zfish | PMID:31594822 |
| Biological Process | GO:0006338 | chromatin remodeling | pou5f3 | IMP | ZFIN:ZDB-GENO-221020-6 | Zfish | PMID:31940339 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | pou5f3 | IMP | ZFIN:ZDB-GENO-070223-1 | Zfish | PMID:16775002 |
| Biological Process | GO:0007398 | ectoderm development | pou5f3 | IMP | ZFIN:ZDB-GENO-020426-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0007398 | ectoderm development | pou5f3 | IMP | ZFIN:ZDB-GENO-020625-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0009880 | embryonic pattern specification | pou5f3 | IMP | ZFIN:ZDB-GENO-020625-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0009880 | embryonic pattern specification | pou5f3 | IMP | ZFIN:ZDB-GENO-100414-8 | Zfish | PMID:15366005 |
| Biological Process | GO:0009880 | embryonic pattern specification | pou5f3 | IMP | ZFIN:ZDB-GENO-020426-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0001706 | endoderm formation | pou5f3 | IMP | ZFIN:ZDB-GENO-020117-1 | Zfish | PMID:14711414 |
| Biological Process | GO:0001706 | endoderm formation | pou5f3 | IMP | ZFIN:ZDB-GENO-020426-1 | Zfish | PMID:14723850 |
| Biological Process | GO:0090504 | epiboly | pou5f3 | IMP | ZFIN:ZDB-GENO-110725-7 | Zfish | PMID:23484854 |
| Biological Process | GO:0055113 | epiboly involved in gastrulation with mouth forming second | pou5f3 | IMP | ZFIN:ZDB-GENO-081023-3 | Zfish | PMID:18215655 |
| Biological Process | GO:0055113 | epiboly involved in gastrulation with mouth forming second | pou5f3 | IMP | ZFIN:ZDB-GENO-081023-1 | Zfish | PMID:18215655 |
| Biological Process | GO:0055113 | epiboly involved in gastrulation with mouth forming second | pou5f3 | IMP | ZFIN:ZDB-GENO-081023-2 | Zfish | PMID:18215655 |
| Biological Process | GO:0031101 | fin regeneration | pou5f3 | IMP | ZFIN:ZDB-MRPHLNO-081015-1 | Zfish | PMID:20089153 |
| Biological Process | GO:0030902 | hindbrain development | pou5f3 | IMP | ZFIN:ZDB-GENO-980202-270 | Zfish | PMID:11923201 |
| Biological Process | GO:0030902 | hindbrain development | pou5f3 | IMP | ZFIN:ZDB-GENO-020117-1 | Zfish | PMID:11923201 |
| Biological Process | GO:0007498 | mesoderm development | pou5f3 | IMP | ZFIN:ZDB-GENO-020426-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0007498 | mesoderm development | pou5f3 | IMP | ZFIN:ZDB-GENO-020625-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0030917 | midbrain-hindbrain boundary development | pou5f3 | IMP | ZFIN:ZDB-GENO-980202-270 | Zfish | PMID:9007238 |
| Biological Process | GO:0016331 | morphogenesis of embryonic epithelium | pou5f3 | IMP | ZFIN:ZDB-GENO-020625-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0016331 | morphogenesis of embryonic epithelium | pou5f3 | IMP | ZFIN:ZDB-GENO-020426-1 | Zfish | PMID:11861475 |
| Biological Process | GO:0034728 | nucleosome organization | pou5f3 | IDA | Zfish | PMID:30674556 | |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | pou5f3 | IGI | ZFIN:ZDB-GENE-011026-1 | Zfish | PMID:14723850 |
| Biological Process | GO:0044030 | regulation of DNA methylation | pou5f3 | IGI | ZFIN:ZDB-GENO-090916-1,ZFIN:ZDB-MRPHLNO-081015-1 | Zfish | PMID:20850014 |
| Biological Process | GO:0042663 | regulation of endodermal cell fate specification | pou5f3 | IGI | ZFIN:ZDB-GENE-010108-1,ZFIN:ZDB-GENE-011026-1 | Zfish | PMID:23364327 |
| Biological Process | GO:0042663 | regulation of endodermal cell fate specification | pou5f3 | IGI | ZFIN:ZDB-GENE-011026-1 | Zfish | PMID:23364327 |
| Biological Process | GO:0010468 | regulation of gene expression | pou5f3 | IMP | ZFIN:ZDB-GENO-110725-7 | Zfish | PMID:24643012 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 07/05/2024 MGI 6.24 |

|
|
|
||