Comparative GO Graph (mouse, human, rat, zebrafish)
Compare experimental GO annotations for Human-Mouse-Rat-Zebrafish orthologs to Mouse Psmc3
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
- This class contains
- Human genes PSMC3
- Mouse genes Psmc3
- Rat genes Psmc3
- Zfish genes psmc3
- Annotations are indicated with nodes colored by organism:
Human,
Mouse,
Rat,
Zfish
- Relations between GO terms are indicated by colored edges "is_a"; "part_of"; "regulates"; "positively_regulates"; "negatively_regulates"
- GO annotations: Human from GOA; Mouse from MGI; Rat from RGD; Zebrafish from ZFIN.
- Only experimental annotations are displayed: EXP, IDA, IEP, IGI, IMP, IPI, HTP, HDA, HMP, HGI, HEP. Evidence codes are listed at the bottom of the page.
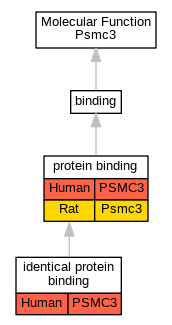
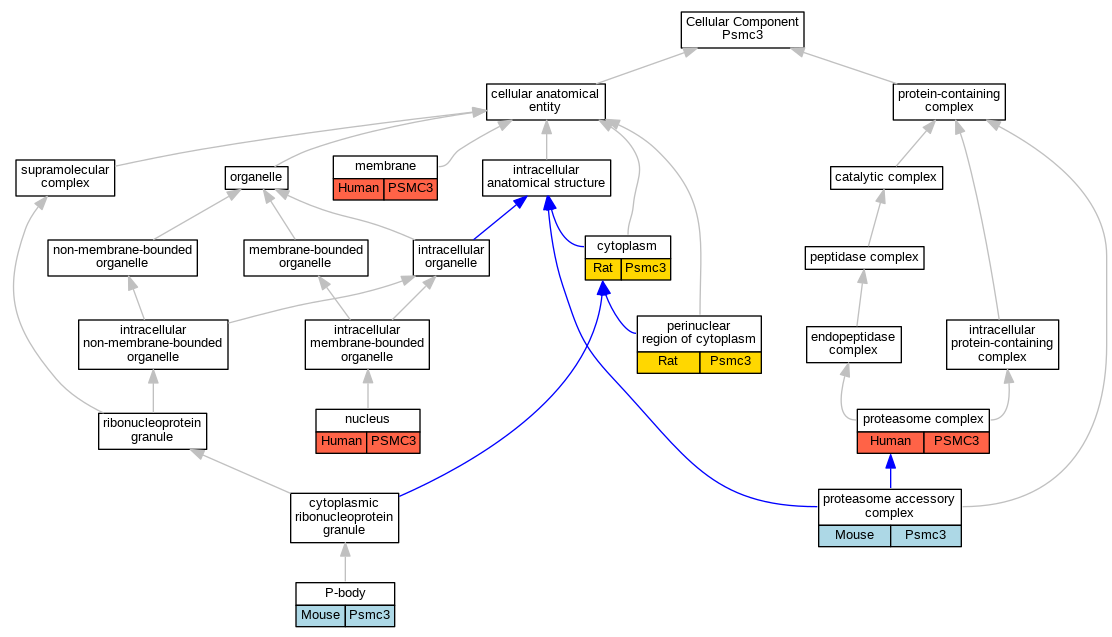
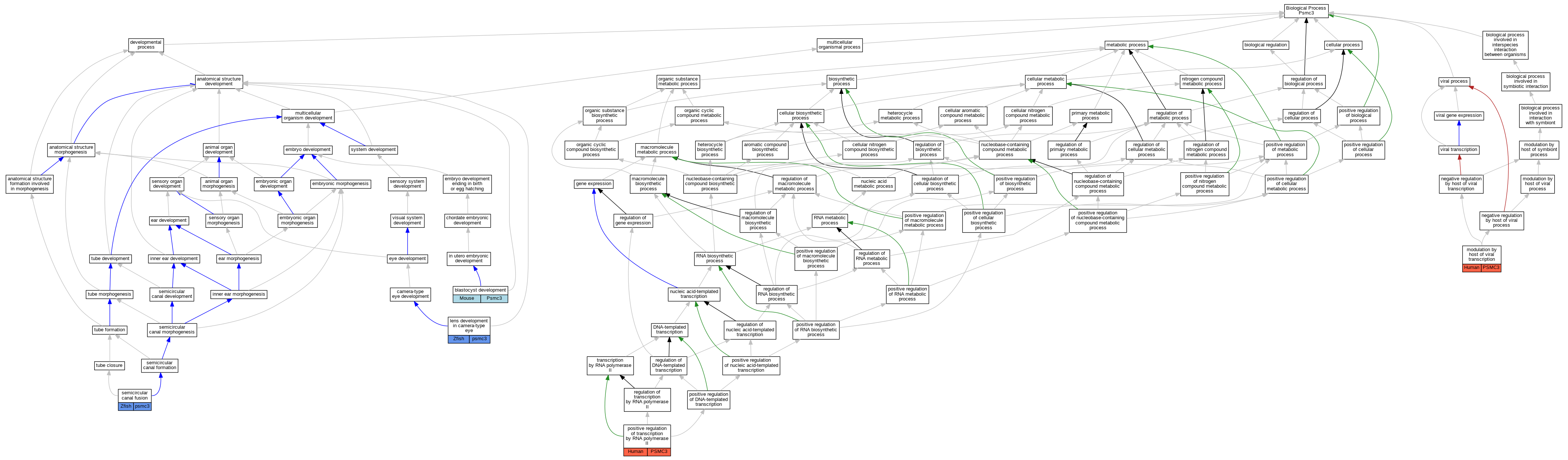 Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Psmc3
Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Psmc3
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0042802 | identical protein binding | PSMC3 | IPI | UniProtKB:P17980 | Human | PMID:25416956 |
| Molecular Function | GO:0042802 | identical protein binding | PSMC3 | IPI | UniProtKB:P17980 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P54253 | Human | PMID:16713569 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P03255-1 | Human | PMID:16763564 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:O00233 | Human | PMID:19490896 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P62333 | Human | PMID:19490896 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:Q9Y2J4 | Human | PMID:24255178 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:A8K2R3 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:O00233 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P62333 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:Q9Y2J4-4 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P62333 | Human | PMID:27342858 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P62333 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:Q9BUZ4 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:Q9NVF7 | Human | PMID:28514442 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P62333 | Human | PMID:29636472 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:O00233 | Human | PMID:31515488 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:O00233 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:O14561 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P12532 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P62333 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:Q9BUZ4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:Q9C005 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:Q9NVF7 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:Q9Y2J4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:O43464 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P00441 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P05067 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P37840 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P42858 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:P54253 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | PSMC3 | IPI | UniProtKB:Q14203-5 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | Psmc3 | IPI | RGD:3729 | Rat | PMID:11032911|RGD:2291798 |
| Molecular Function | GO:0005515 | protein binding | Psmc3 | IPI | UniProtKB:A0A0G2K916,UniProtKB:B2GV38,UniProtKB:B5DFN4,UniProtKB:D3Z941,UniProtKB:D4AA63,UniProtKB:F1M853,UniProtKB:P11250,UniProtKB:P51593,UniProtKB:P61107,UniProtKB:P62718,UniProtKB:P97612,UniProtKB:Q09167,UniProtKB:Q498M4,UniProtKB:Q5PPJ6,UniProtKB:Q5PPK9,UniProtKB:Q6AYJ7 | Rat | PMID:27191843|RGD:11344934 |
| Cellular Component | GO:0005737 | cytoplasm | Psmc3 | IDA | | Rat | PMID:11032911|RGD:2291798 |
| Cellular Component | GO:0016020 | membrane | PSMC3 | HDA | | Human | PMID:19946888 |
| Cellular Component | GO:0005634 | nucleus | PSMC3 | HDA | | Human | PMID:21630459 |
| Cellular Component | GO:0005634 | nucleus | PSMC3 | IDA | | Human | PMID:2194290 |
| Cellular Component | GO:0000932 | P-body | Psmc3 | IDA | | Mouse | MGI:MGI:5487513|PMID:22078707 |
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | Psmc3 | IDA | | Rat | PMID:11032911|RGD:2291798 |
| Cellular Component | GO:0022624 | proteasome accessory complex | Psmc3 | IDA | | Mouse | MGI:MGI:5442509|PMID:16857966 |
| Cellular Component | GO:0000502 | proteasome complex | PSMC3 | IDA | | Human | PMID:17323924 |
| Cellular Component | GO:0000502 | proteasome complex | PSMC3 | IDA | | Human | PMID:9464850 |
| Biological Process | GO:0001824 | blastocyst development | Psmc3 | IMP | MGI:MGI:2180548 | Mouse | PMID:10945464 |
| Biological Process | GO:0002088 | lens development in camera-type eye | psmc3 | IMP | ZFIN:ZDB-CRISPR-210222-3 | Zfish | PMID:32500975 |
| Biological Process | GO:0002088 | lens development in camera-type eye | psmc3 | IMP | ZFIN:ZDB-MRPHLNO-210222-2 | Zfish | PMID:32500975 |
| Biological Process | GO:0002088 | lens development in camera-type eye | psmc3 | IMP | ZFIN:ZDB-CRISPR-210222-4 | Zfish | PMID:32500975 |
| Biological Process | GO:0043921 | modulation by host of viral transcription | PSMC3 | IDA | | Human | PMID:2194290 |
| Biological Process | GO:0043921 | modulation by host of viral transcription | PSMC3 | IDA | | Human | PMID:8419915 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | PSMC3 | IDA | | Human | PMID:19853614 |
| Biological Process | GO:0060879 | semicircular canal fusion | psmc3 | IMP | ZFIN:ZDB-CRISPR-210222-3 | Zfish | PMID:32500975 |
| Biological Process | GO:0060879 | semicircular canal fusion | psmc3 | IMP | ZFIN:ZDB-MRPHLNO-210222-2 | Zfish | PMID:32500975 |
| Biological Process | GO:0060879 | semicircular canal fusion | psmc3 | IMP | ZFIN:ZDB-CRISPR-210222-4 | Zfish | PMID:32500975 |
Gene Ontology Evidence Code Abbreviations:
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools





