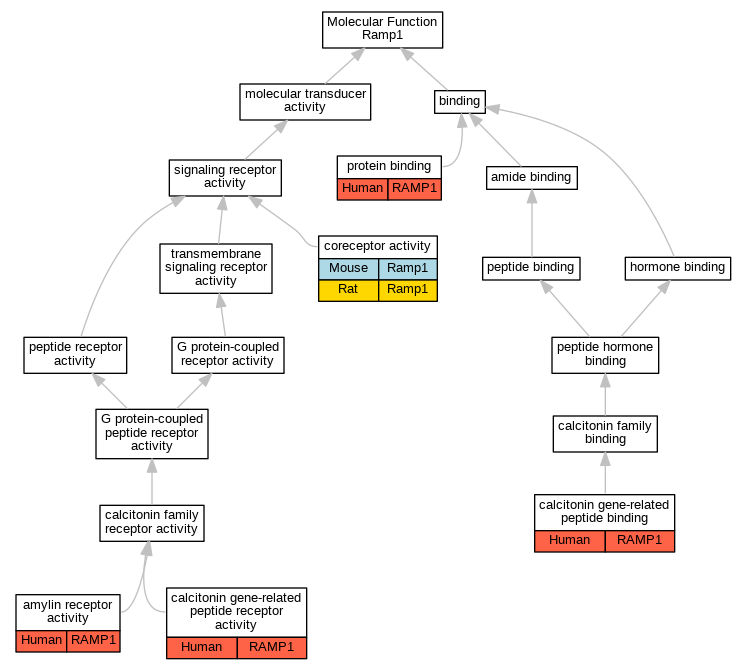
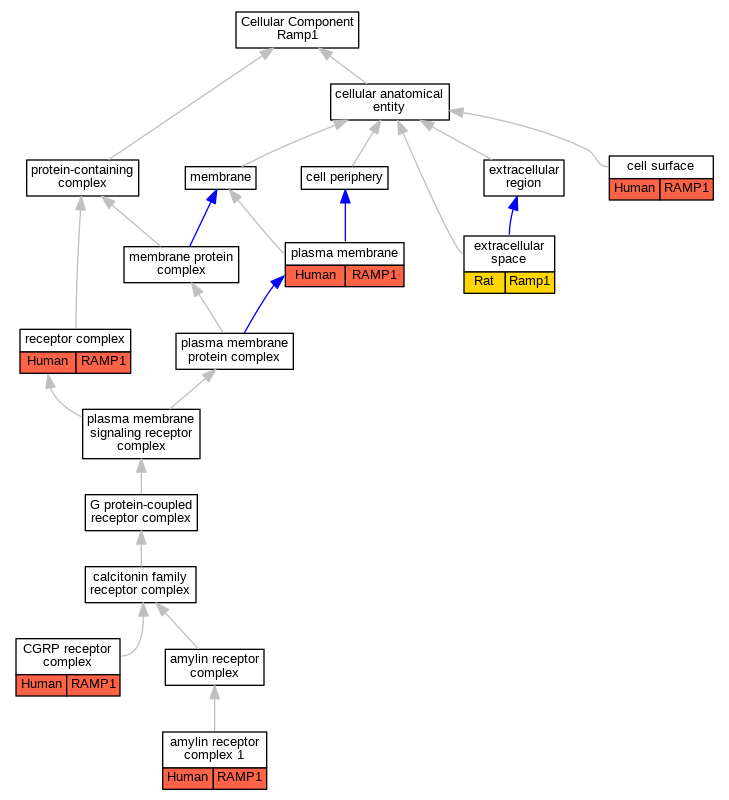
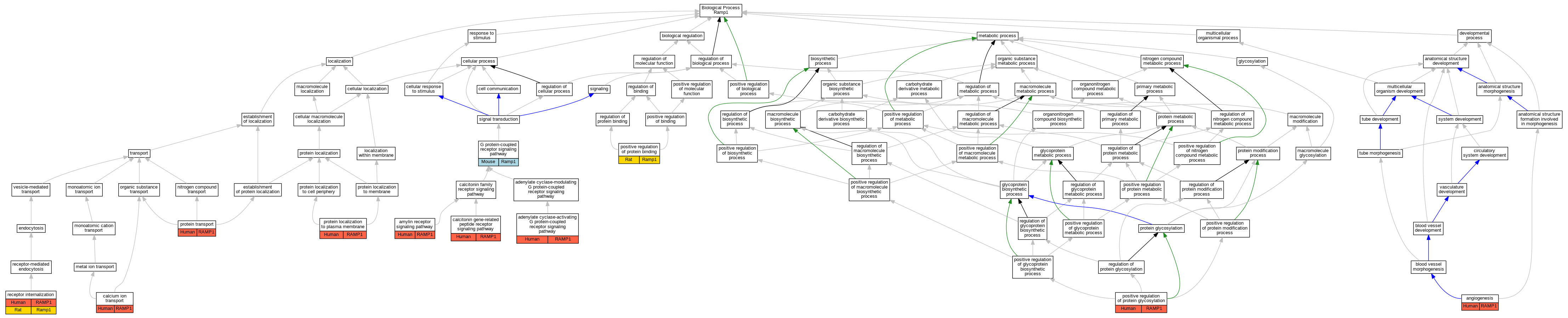
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0097643 | amylin receptor activity | RAMP1 | IGI | UniProtKB:P06881,UniProtKB:P30988|UniProtKB:P10997:PRO_0000004106,UniProtKB:P30988|UniProtKB:P12969:PRO_0000004122,UniProtKB:P30988|UniProtKB:P30988,UniProtKB:P35318 | Human | PMID:22500019 |
| Molecular Function | GO:0097643 | amylin receptor activity | RAMP1 | IPI | UniProtKB:P30988 | Human | PMID:14722252 |
| Molecular Function | GO:1990407 | calcitonin gene-related peptide binding | RAMP1 | IPI | UniProtKB:Q16602 | Human | PMID:9620797 |
| Molecular Function | GO:0001635 | calcitonin gene-related peptide receptor activity | RAMP1 | IPI | UniProtKB:Q16602 | Human | PMID:10882736 |
| Molecular Function | GO:0001635 | calcitonin gene-related peptide receptor activity | RAMP1 | IPI | UniProtKB:P30988 | Human | PMID:14722252 |
| Molecular Function | GO:0001635 | calcitonin gene-related peptide receptor activity | RAMP1 | IPI | UniProtKB:Q16602 | Human | PMID:9620797 |
| Molecular Function | GO:0015026 | coreceptor activity | Ramp1 | IDA | | Mouse | PMID:10854696 |
| Molecular Function | GO:0015026 | coreceptor activity | Ramp1 | IDA | | Rat | PMID:11847213|RGD:1299114 |
| Molecular Function | GO:0015026 | coreceptor activity | Ramp1 | IMP | | Rat | PMID:16242145|RGD:1625358 |
| Molecular Function | GO:0005515 | protein binding | RAMP1 | IPI | UniProtKB:P06881 | Human | PMID:14722252 |
| Molecular Function | GO:0005515 | protein binding | RAMP1 | IPI | UniProtKB:Q16602 | Human | PMID:16293613 |
| Molecular Function | GO:0005515 | protein binding | RAMP1 | IPI | UniProtKB:Q16602 | Human | PMID:20596610 |
| Molecular Function | GO:0005515 | protein binding | RAMP1 | IPI | UniProtKB:Q16602 | Human | PMID:20826335 |
| Molecular Function | GO:0005515 | protein binding | RAMP1 | IPI | UniProtKB:P21145 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | RAMP1 | IPI | UniProtKB:Q16617 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | RAMP1 | IPI | UniProtKB:Q5J8X5 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | RAMP1 | IPI | UniProtKB:Q16602 | Human | PMID:33602864 |
| Molecular Function | GO:0005515 | protein binding | RAMP1 | IPI | UniProtKB:P06881 | Human | PMID:9620797 |
| Cellular Component | GO:0150056 | amylin receptor complex 1 | RAMP1 | IDA | | Human | PMID:22500019 |
| Cellular Component | GO:0009986 | cell surface | RAMP1 | IDA | | Human | PMID:14722252 |
| Cellular Component | GO:1990406 | CGRP receptor complex | RAMP1 | IDA | | Human | PMID:10882736 |
| Cellular Component | GO:1990406 | CGRP receptor complex | RAMP1 | IDA | | Human | PMID:9620797 |
| Cellular Component | GO:0005615 | extracellular space | Ramp1 | IDA | | Rat | PMID:11847213|RGD:1299114 |
| Cellular Component | GO:0005886 | plasma membrane | RAMP1 | IDA | | Human | PMID:9620797 |
| Cellular Component | GO:0005886 | plasma membrane | RAMP1 | IDA | | Human | PMID:18039931 |
| Cellular Component | GO:0043235 | receptor complex | RAMP1 | IDA | | Human | PMID:15613468 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | RAMP1 | IPI | UniProtKB:Q16602 | Human | PMID:10882736 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | RAMP1 | IDA | | Human | PMID:14722252 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | RAMP1 | IGI | UniProtKB:P06881,UniProtKB:P30988 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | RAMP1 | IGI | UniProtKB:P10997:PRO_0000004106,UniProtKB:P30988 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | RAMP1 | IGI | UniProtKB:P12969:PRO_0000004122,UniProtKB:P30988 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | RAMP1 | IGI | UniProtKB:P30988,UniProtKB:P35318 | Human | PMID:22500019 |
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | RAMP1 | IPI | UniProtKB:Q16602 | Human | PMID:9620797 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | RAMP1 | IDA | | Human | PMID:14722252 |
| Biological Process | GO:0097647 | amylin receptor signaling pathway | RAMP1 | IGI | UniProtKB:P06881,UniProtKB:P30988|UniProtKB:P10997:PRO_0000004106,UniProtKB:P30988|UniProtKB:P12969:PRO_0000004122,UniProtKB:P30988|UniProtKB:P30988,UniProtKB:P35318 | Human | PMID:22500019 |
| Biological Process | GO:0001525 | angiogenesis | RAMP1 | IDA | | Human | PMID:20596610 |
| Biological Process | GO:1990408 | calcitonin gene-related peptide receptor signaling pathway | RAMP1 | IPI | UniProtKB:Q16602 | Human | PMID:10882736 |
| Biological Process | GO:1990408 | calcitonin gene-related peptide receptor signaling pathway | RAMP1 | IPI | UniProtKB:Q16602 | Human | PMID:9620797 |
| Biological Process | GO:0006816 | calcium ion transport | RAMP1 | IDA | | Human | PMID:10882736 |
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | Ramp1 | IPI | UniProtKB:P70160 | Mouse | PMID:10854696 |
| Biological Process | GO:0032092 | positive regulation of protein binding | Ramp1 | IDA | | Rat | PMID:11556887|RGD:1299115 |
| Biological Process | GO:0060050 | positive regulation of protein glycosylation | RAMP1 | IDA | | Human | PMID:9620797 |
| Biological Process | GO:0072659 | protein localization to plasma membrane | RAMP1 | IDA | | Human | PMID:10882736 |
| Biological Process | GO:0015031 | protein transport | RAMP1 | IDA | | Human | PMID:10882736 |
| Biological Process | GO:0031623 | receptor internalization | RAMP1 | IDA | | Human | PMID:10882736 |
| Biological Process | GO:0031623 | receptor internalization | RAMP1 | IDA | | Human | PMID:15613468 |
| Biological Process | GO:0031623 | receptor internalization | Ramp1 | IDA | | Rat | PMID:15613468|RGD:8554018 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools