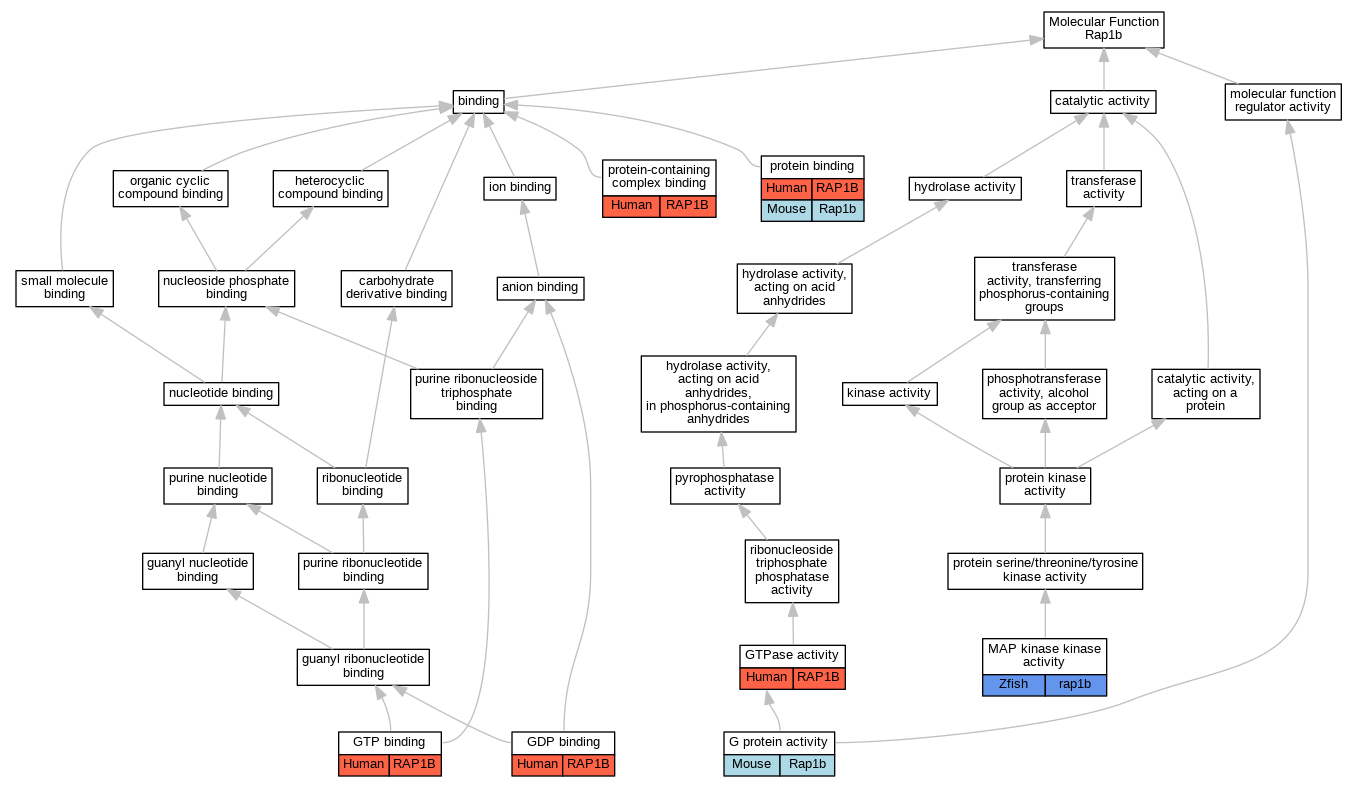
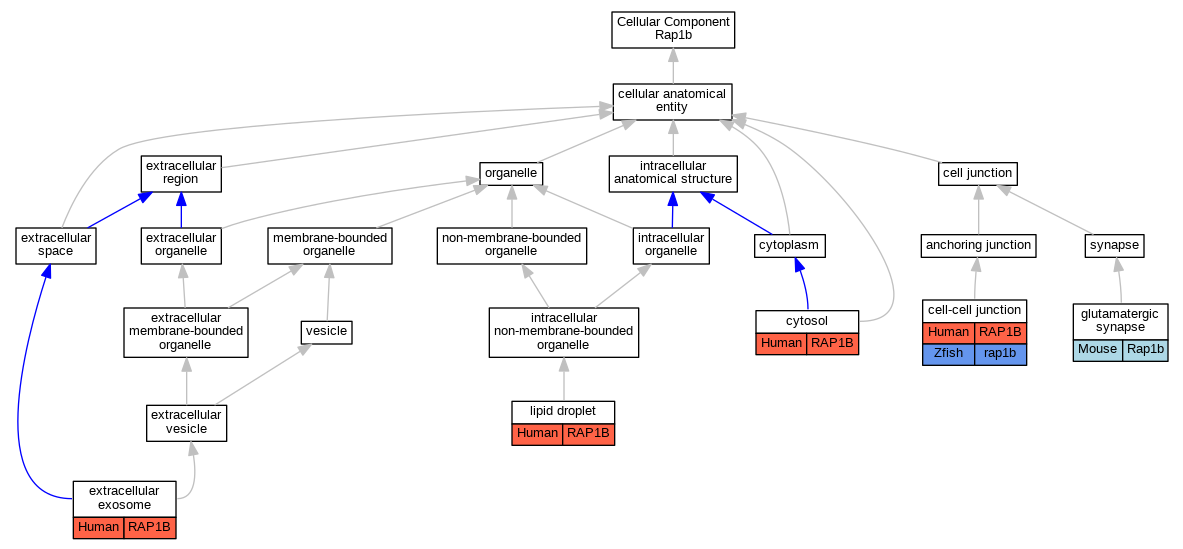
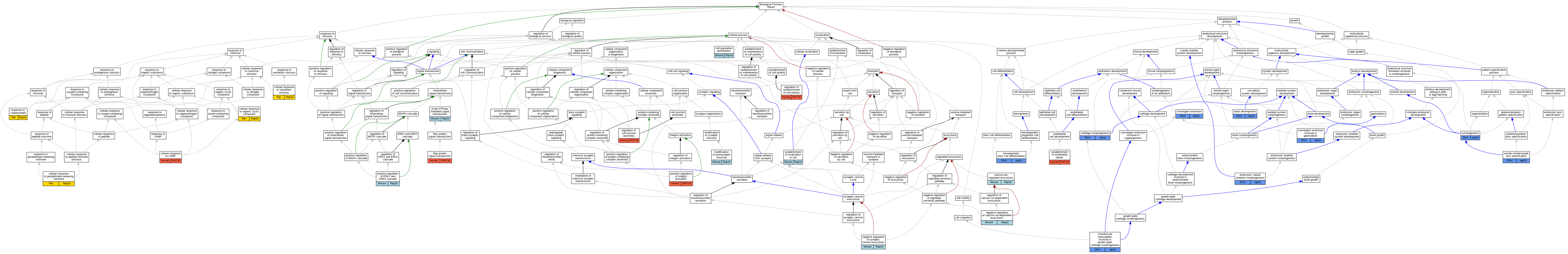
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0003925 | G protein activity | Rap1b | IDA | | Mouse | PMID:11056535 |
| Molecular Function | GO:0019003 | GDP binding | RAP1B | IDA | | Human | PMID:18309292 |
| Molecular Function | GO:0005525 | GTP binding | RAP1B | IDA | | Human | PMID:18309292 |
| Molecular Function | GO:0003924 | GTPase activity | RAP1B | IDA | | Human | PMID:18309292 |
| Molecular Function | GO:0004708 | MAP kinase kinase activity | rap1b | IGI | ZFIN:ZDB-MRPHLNO-070519-1,ZFIN:ZDB-MRPHLNO-070519-2 | Zfish | PMID:26280580 |
| Molecular Function | GO:0004708 | MAP kinase kinase activity | rap1b | IGI | ZFIN:ZDB-MRPHLNO-070519-1,ZFIN:ZDB-MRPHLNO-070519-2,ZFIN:ZDB-MRPHLNO-151208-5 | Zfish | PMID:26280580 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:Q9EQZ6-3 | Human | PMID:16452984 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:P47736 | Human | PMID:18309292 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:Q9EQZ6 | Human | PMID:18660803 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:Q9EQZ6-3 | Human | PMID:18660803 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:Q12967 | Human | PMID:21988832 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:Q12967 | Human | PMID:22705156 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:P52306 | Human | PMID:24415755 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:Q8WWW0 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:Q5U651 | Human | PMID:26780829 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:Q12967-6 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:Q5JR59-3 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:P27797 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:P36957 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | RAP1B | IPI | UniProtKB:Q8TDX7 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | Rap1b | IPI | PR:Q99N57 | Mouse | PMID:14712229 |
| Molecular Function | GO:0044877 | protein-containing complex binding | RAP1B | IDA | | Human | PMID:23209302 |
| Cellular Component | GO:0005911 | cell-cell junction | RAP1B | IDA | | Human | PMID:20332120 |
| Cellular Component | GO:0005911 | cell-cell junction | rap1b | IMP | ZFIN:ZDB-MRPHLNO-090826-11 | Zfish | PMID:19093037 |
| Cellular Component | GO:0005829 | cytosol | RAP1B | IDA | | Human | PMID:18582561 |
| Cellular Component | GO:0070062 | extracellular exosome | RAP1B | HDA | | Human | PMID:19199708 |
| Cellular Component | GO:0070062 | extracellular exosome | RAP1B | HDA | | Human | PMID:20458337 |
| Cellular Component | GO:0098978 | glutamatergic synapse | Rap1b | IMP | | Mouse | PMID:26844834 |
| Cellular Component | GO:0098978 | glutamatergic synapse | Rap1b | IDA | | Mouse | PMID:26844834 |
| Cellular Component | GO:0005811 | lipid droplet | RAP1B | IDA | | Human | PMID:14741744 |
| Biological Process | GO:0017156 | calcium-ion regulated exocytosis | Rap1b | IGI | MGI:MGI:97852 | Mouse | PMID:23616533 |
| Biological Process | GO:0060536 | cartilage morphogenesis | rap1b | IGI | ZFIN:ZDB-MRPHLNO-070519-1,ZFIN:ZDB-MRPHLNO-070519-2 | Zfish | PMID:26280580 |
| Biological Process | GO:0008283 | cell population proliferation | Rap1b | IPI | UniProtKB:Q91ZZ2 | Mouse | PMID:14712229 |
| Biological Process | GO:0008283 | cell population proliferation | Rap1b | IPI | UniProtKB:Q8VCC8 | Mouse | PMID:14712229 |
| Biological Process | GO:0071320 | cellular response to cAMP | RAP1B | IDA | | Human | PMID:21840392 |
| Biological Process | GO:0097211 | cellular response to gonadotropin-releasing hormone | Rap1b | IEP | | Rat | PMID:22127306|RGD:10041018 |
| Biological Process | GO:0071407 | cellular response to organic cyclic compound | Rap1b | IDA | | Rat | PMID:10908723|RGD:10003157 |
| Biological Process | GO:0071466 | cellular response to xenobiotic stimulus | Rap1b | IDA | | Rat | PMID:10908723|RGD:10003157 |
| Biological Process | GO:0003428 | chondrocyte intercalation involved in growth plate cartilage morphogenesis | rap1b | IGI | ZFIN:ZDB-MRPHLNO-070519-1,ZFIN:ZDB-MRPHLNO-070519-2 | Zfish | PMID:26280580 |
| Biological Process | GO:0060026 | convergent extension | rap1b | IGI | ZFIN:ZDB-MRPHLNO-070519-1,ZFIN:ZDB-MRPHLNO-070519-2 | Zfish | PMID:26280580 |
| Biological Process | GO:0060026 | convergent extension | rap1b | IMP | ZFIN:ZDB-MRPHLNO-070519-3 | Zfish | PMID:17336901 |
| Biological Process | GO:0060026 | convergent extension | rap1b | IGI | ZFIN:ZDB-CRISPR-151208-1,ZFIN:ZDB-CRISPR-151208-2 | Zfish | PMID:26280580 |
| Biological Process | GO:0060026 | convergent extension | rap1b | IGI | ZFIN:ZDB-MRPHLNO-070519-1,ZFIN:ZDB-MRPHLNO-070519-2,ZFIN:ZDB-MRPHLNO-151208-3 | Zfish | PMID:26280580 |
| Biological Process | GO:0060026 | convergent extension | rap1b | IMP | ZFIN:ZDB-MRPHLNO-070519-2 | Zfish | PMID:17336901 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | rap1b | IGI | ZFIN:ZDB-CRISPR-151208-1,ZFIN:ZDB-CRISPR-151208-2 | Zfish | PMID:30018450 |
| Biological Process | GO:0048701 | embryonic cranial skeleton morphogenesis | rap1b | IGI | ZFIN:ZDB-CRISPR-151208-1,ZFIN:ZDB-CRISPR-151208-2 | Zfish | PMID:30018450 |
| Biological Process | GO:0061028 | establishment of endothelial barrier | RAP1B | IMP | | Human | PMID:21840392 |
| Biological Process | GO:0051649 | establishment of localization in cell | Rap1b | IGI | MGI:MGI:97852 | Mouse | PMID:23616533 |
| Biological Process | GO:0007507 | heart development | rap1b | IGI | ZFIN:ZDB-MRPHLNO-070519-1,ZFIN:ZDB-MRPHLNO-070519-3 | Zfish | PMID:23226434 |
| Biological Process | GO:0060218 | hematopoietic stem cell differentiation | rap1b | IMP | ZFIN:ZDB-GENO-200409-12 | Zfish | PMID:31006651 |
| Biological Process | GO:0099010 | modification of postsynaptic structure | Rap1b | IDA | | Mouse | PMID:26844834 |
| Biological Process | GO:0099010 | modification of postsynaptic structure | Rap1b | IMP | | Mouse | PMID:26844834 |
| Biological Process | GO:0045955 | negative regulation of calcium ion-dependent exocytosis | Rap1b | IGI | MGI:MGI:97852 | Mouse | PMID:23616533 |
| Biological Process | GO:2000301 | negative regulation of synaptic vesicle exocytosis | Rap1b | IGI | MGI:MGI:97852 | Mouse | PMID:23616533 |
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | Rap1b | IGI | MGI:MGI:97852 | Mouse | PMID:23616533 |
| Biological Process | GO:0033625 | positive regulation of integrin activation | RAP1B | IMP | | Human | PMID:19461049 |
| Biological Process | GO:0032486 | Rap protein signal transduction | RAP1B | IMP | | Human | PMID:21840392 |
| Biological Process | GO:1901888 | regulation of cell junction assembly | RAP1B | IMP | | Human | PMID:21840392 |
| Biological Process | GO:2000114 | regulation of establishment of cell polarity | RAP1B | IMP | | Human | PMID:20332120 |
| Biological Process | GO:0009743 | response to carbohydrate | Rap1b | IEP | | Rat | PMID:9149109|RGD:633760 |
| Biological Process | GO:0007264 | small GTPase mediated signal transduction | Rap1b | IDA | | Mouse | PMID:14712229 |
| Biological Process | GO:0032525 | somite rostral/caudal axis specification | rap1b | IGI | ZFIN:ZDB-GENO-100512-3,ZFIN:ZDB-MRPHLNO-070519-3 | Zfish | PMID:23192979 |
| Biological Process | GO:0032525 | somite rostral/caudal axis specification | rap1b | IGI | ZFIN:ZDB-GENO-100512-2,ZFIN:ZDB-MRPHLNO-070519-3 | Zfish | PMID:23192979 |
| Biological Process | GO:0001756 | somitogenesis | rap1b | IGI | ZFIN:ZDB-GENO-100512-2,ZFIN:ZDB-MRPHLNO-070519-3 | Zfish | PMID:23192979 |
| Biological Process | GO:0001756 | somitogenesis | rap1b | IGI | ZFIN:ZDB-GENO-100512-3,ZFIN:ZDB-MRPHLNO-070519-3 | Zfish | PMID:23192979 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools