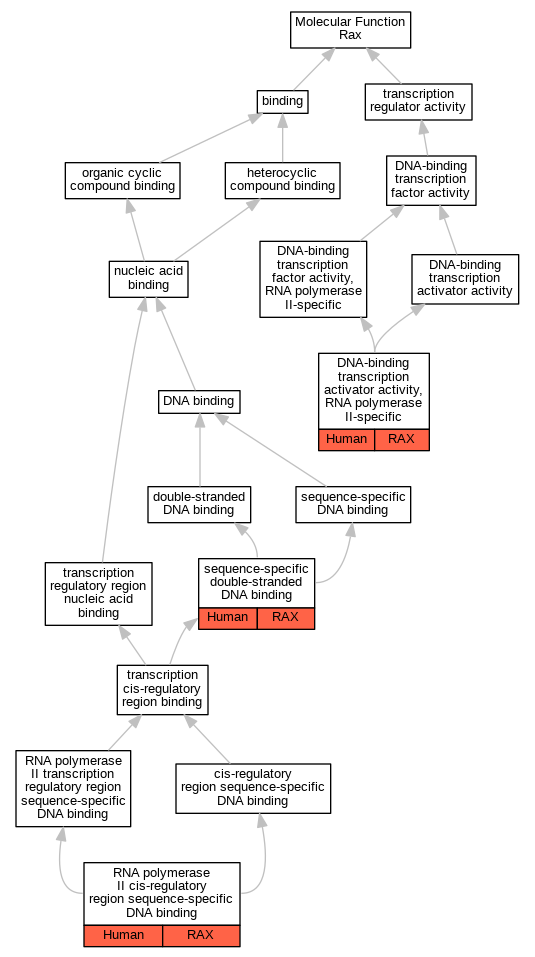
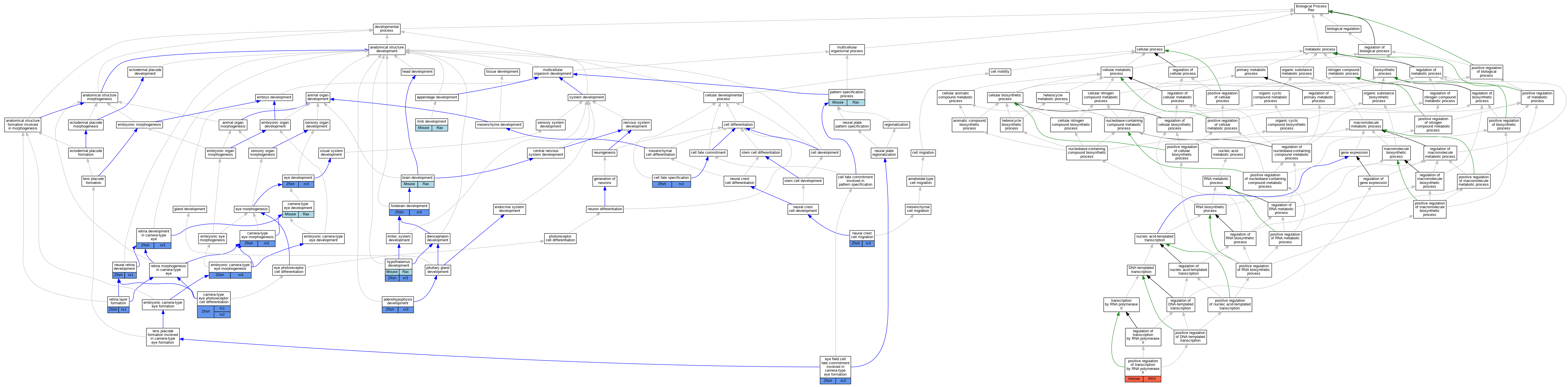
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | RAX | IDA | | Human | PMID:10625658 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | RAX | IDA | | Human | PMID:10625658 |
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | RAX | IDA | | Human | PMID:28473536 |
| Biological Process | GO:0021984 | adenohypophysis development | rx3 | IMP | ZFIN:ZDB-GENO-051229-1 | Zfish | PMID:17373855 |
| Biological Process | GO:0007420 | brain development | Rax | IMP | MGI:MGI:2152531 | Mouse | PMID:15789424 |
| Biological Process | GO:0043010 | camera-type eye development | Rax | IMP | MGI:MGI:2152531 | Mouse | PMID:11105055 |
| Biological Process | GO:0043010 | camera-type eye development | Rax | IMP | MGI:MGI:1856862 | Mouse | PMID:17247120 |
| Biological Process | GO:0043010 | camera-type eye development | Rax | IMP | MGI:MGI:2152531 | Mouse | PMID:15789424 |
| Biological Process | GO:0043010 | camera-type eye development | Rax | IMP | MGI:MGI:1856862 | Mouse | PMID:640224 |
| Biological Process | GO:0043010 | camera-type eye development | Rax | IMP | MGI:MGI:1856862 | Mouse | PMID:13970081 |
| Biological Process | GO:0048593 | camera-type eye morphogenesis | rx3 | IMP | ZFIN:ZDB-GENO-081016-1 | Zfish | PMID:12947416 |
| Biological Process | GO:0060219 | camera-type eye photoreceptor cell differentiation | rx1 | IGI | ZFIN:ZDB-MRPHLNO-060109-2,ZFIN:ZDB-MRPHLNO-060109-4,ZFIN:ZDB-MRPHLNO-100423-2,ZFIN:ZDB-MRPHLNO-100423-3 | Zfish | PMID:19210961 |
| Biological Process | GO:0060219 | camera-type eye photoreceptor cell differentiation | rx1 | IMP | ZFIN:ZDB-MRPHLNO-060109-2,ZFIN:ZDB-MRPHLNO-100423-2 | Zfish | PMID:19210961 |
| Biological Process | GO:0060219 | camera-type eye photoreceptor cell differentiation | rx2 | IGI | ZFIN:ZDB-MRPHLNO-060109-2,ZFIN:ZDB-MRPHLNO-060109-4,ZFIN:ZDB-MRPHLNO-100423-2,ZFIN:ZDB-MRPHLNO-100423-3 | Zfish | PMID:19210961 |
| Biological Process | GO:0001708 | cell fate specification | rx3 | IMP | ZFIN:ZDB-GENO-051229-2 | Zfish | PMID:16300752 |
| Biological Process | GO:0001708 | cell fate specification | rx3 | IMP | ZFIN:ZDB-GENO-051229-1 | Zfish | PMID:16300752 |
| Biological Process | GO:0048596 | embryonic camera-type eye morphogenesis | rx3 | IMP | ZFIN:ZDB-GENO-051229-2 | Zfish | PMID:16300752 |
| Biological Process | GO:0048596 | embryonic camera-type eye morphogenesis | rx3 | IMP | ZFIN:ZDB-MRPHLNO-060109-5 | Zfish | PMID:16300752 |
| Biological Process | GO:0048596 | embryonic camera-type eye morphogenesis | rx3 | IMP | ZFIN:ZDB-GENO-051229-1 | Zfish | PMID:16300752 |
| Biological Process | GO:0048596 | embryonic camera-type eye morphogenesis | rx3 | IMP | ZFIN:ZDB-GENO-060822-1 | Zfish | PMID:16818451 |
| Biological Process | GO:0048596 | embryonic camera-type eye morphogenesis | rx3 | IMP | ZFIN:ZDB-MRPHLNO-060109-6 | Zfish | PMID:16300752 |
| Biological Process | GO:0001654 | eye development | rx3 | IMP | ZFIN:ZDB-GENO-040513-1 | Zfish | PMID:15183718 |
| Biological Process | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation | rx3 | IMP | ZFIN:ZDB-GENO-131219-2 | Zfish | PMID:24026122 |
| Biological Process | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation | rx3 | IMP | ZFIN:ZDB-GENO-150212-4 | Zfish | PMID:24026122 |
| Biological Process | GO:0030900 | forebrain development | rx3 | IMP | ZFIN:ZDB-GENO-060822-1 | Zfish | PMID:16818451 |
| Biological Process | GO:0021854 | hypothalamus development | Rax | IMP | MGI:MGI:1856862 | Mouse | PMID:925203 |
| Biological Process | GO:0021854 | hypothalamus development | rx3 | IMP | ZFIN:ZDB-GENO-150206-3 | Zfish | PMID:27317806 |
| Biological Process | GO:0060173 | limb development | Rax | IMP | MGI:MGI:1856862 | Mouse | PMID:13970082 |
| Biological Process | GO:0001755 | neural crest cell migration | rx3 | IMP | ZFIN:ZDB-GENO-081016-1 | Zfish | PMID:18498099 |
| Biological Process | GO:0003407 | neural retina development | rx1 | IMP | ZFIN:ZDB-MRPHLNO-060109-2,ZFIN:ZDB-MRPHLNO-100423-2 | Zfish | PMID:19210961 |
| Biological Process | GO:0007389 | pattern specification process | Rax | IMP | MGI:MGI:1856862 | Mouse | PMID:13970082 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | RAX | IDA | | Human | PMID:10625658 |
| Biological Process | GO:0060041 | retina development in camera-type eye | rx3 | IMP | ZFIN:ZDB-GENO-081016-1 | Zfish | PMID:12947416 |
| Biological Process | GO:0010842 | retina layer formation | rx1 | IMP | ZFIN:ZDB-MRPHLNO-060109-2,ZFIN:ZDB-MRPHLNO-100423-2 | Zfish | PMID:19210961 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools