| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
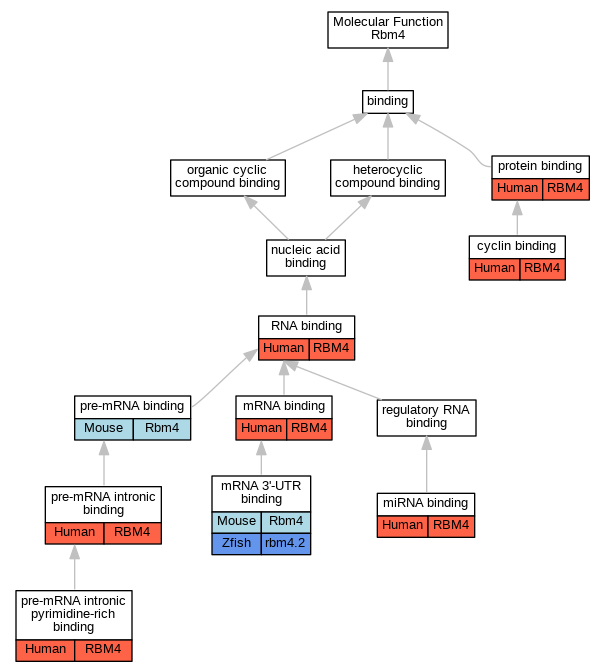
| Molecular Function | GO:0030332 | cyclin binding | RBM4 | IPI | UniProtKB:P78396 | Human | PMID:15159402 |
| Molecular Function | GO:0035198 | miRNA binding | RBM4 | IDA | | Human | PMID:19801630 |
| Molecular Function | GO:0003730 | mRNA 3'-UTR binding | Rbm4 | IDA | | Mouse | PMID:17264215 |
| Molecular Function | GO:0003730 | mRNA 3'-UTR binding | rbm4.2 | IDA | | Zfish | PMID:31227602 |
| Molecular Function | GO:0003729 | mRNA binding | RBM4 | IDA | | Human | PMID:21518792 |
| Molecular Function | GO:0036002 | pre-mRNA binding | Rbm4 | IDA | | Mouse | MGI:MGI:5447617|PMID:16260624 |
| Molecular Function | GO:0097157 | pre-mRNA intronic binding | RBM4 | IDA | | Human | PMID:16260624 |
| Molecular Function | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding | RBM4 | IDA | | Human | PMID:16260624 |
| Molecular Function | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding | RBM4 | IDA | | Human | PMID:16777844 |
| Molecular Function | GO:0005515 | protein binding | RBM4 | IPI | UniProtKB:Q9Y5L0 | Human | PMID:12628928 |
| Molecular Function | GO:0005515 | protein binding | RBM4 | IPI | UniProtKB:P19544 | Human | PMID:16934801 |
| Molecular Function | GO:0005515 | protein binding | RBM4 | IPI | UniProtKB:P60842 | Human | PMID:17284590 |
| Molecular Function | GO:0005515 | protein binding | RBM4 | IPI | UniProtKB:Q04637 | Human | PMID:17284590 |
| Molecular Function | GO:0005515 | protein binding | RBM4 | IPI | UniProtKB:Q9UKV8 | Human | PMID:19801630 |
| Molecular Function | GO:0005515 | protein binding | RBM4 | IPI | UniProtKB:P17844 | Human | PMID:21343338 |
| Molecular Function | GO:0005515 | protein binding | RBM4 | IPI | UniProtKB:P02489 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | RBM4 | IPI | UniProtKB:P22607 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | RBM4 | IPI | UniProtKB:Q86V38 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | RBM4 | IPI | UniProtKB:Q92876 | Human | PMID:32814053 |
| Molecular Function | GO:0003723 | RNA binding | RBM4 | IDA | | Human | PMID:19561594 |
| Molecular Function | GO:0003723 | RNA binding | RBM4 | HDA | | Human | PMID:22658674 |
| Molecular Function | GO:0003723 | RNA binding | RBM4 | HDA | | Human | PMID:22681889 |
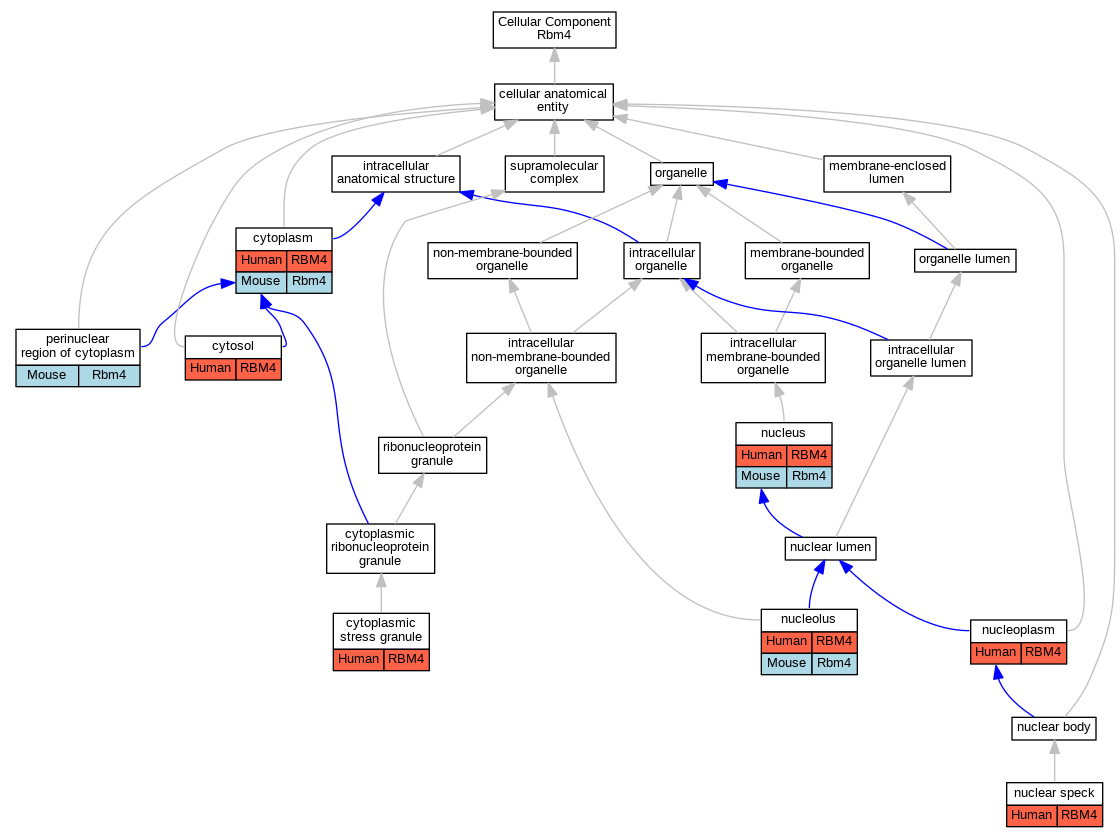
| Cellular Component | GO:0005737 | cytoplasm | RBM4 | IDA | | Human | PMID:12628928 |
| Cellular Component | GO:0005737 | cytoplasm | RBM4 | IDA | | Human | PMID:16777844 |
| Cellular Component | GO:0005737 | cytoplasm | RBM4 | IDA | | Human | PMID:16907643 |
| Cellular Component | GO:0005737 | cytoplasm | RBM4 | IDA | | Human | PMID:17284590 |
| Cellular Component | GO:0005737 | cytoplasm | Rbm4 | IDA | | Mouse | MGI:MGI:4436983|PMID:19801630 |
| Cellular Component | GO:0010494 | cytoplasmic stress granule | RBM4 | IDA | | Human | PMID:17284590 |
| Cellular Component | GO:0005829 | cytosol | RBM4 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0016607 | nuclear speck | RBM4 | IDA | | Human | PMID:12628928 |
| Cellular Component | GO:0016607 | nuclear speck | RBM4 | IDA | | Human | PMID:16907643 |
| Cellular Component | GO:0005730 | nucleolus | RBM4 | IDA | | Human | PMID:12628928 |
| Cellular Component | GO:0005730 | nucleolus | RBM4 | IDA | | Human | PMID:16907643 |
| Cellular Component | GO:0005730 | nucleolus | Rbm4 | IDA | | Mouse | MGI:MGI:4436983|PMID:19801630 |
| Cellular Component | GO:0005654 | nucleoplasm | RBM4 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005654 | nucleoplasm | RBM4 | IDA | | Human | PMID:12628928 |
| Cellular Component | GO:0005634 | nucleus | RBM4 | IDA | | Human | PMID:16777844 |
| Cellular Component | GO:0005634 | nucleus | RBM4 | IDA | | Human | PMID:16907643 |
| Cellular Component | GO:0005634 | nucleus | RBM4 | IDA | | Human | PMID:17284590 |
| Cellular Component | GO:0005634 | nucleus | Rbm4 | IDA | | Mouse | MGI:MGI:4436983|PMID:19801630 |
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | Rbm4 | IDA | | Mouse | MGI:MGI:4436983|PMID:19801630 |
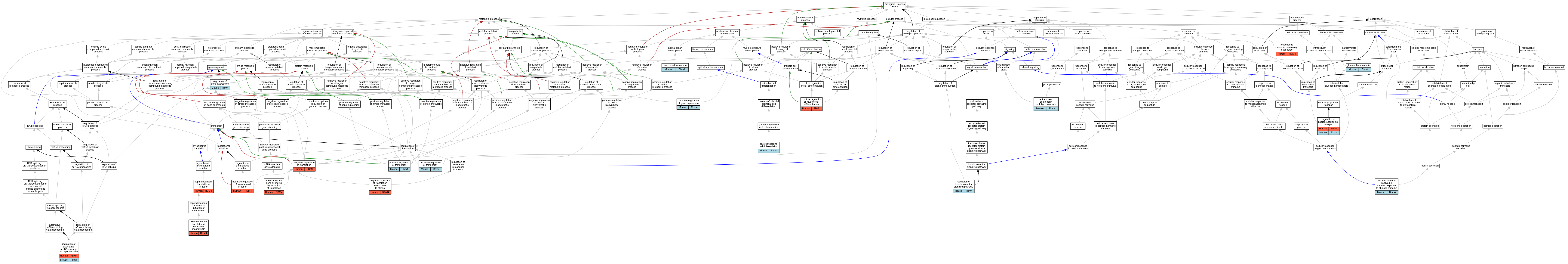
| Biological Process | GO:0002190 | cap-independent translational initiation | RBM4 | IDA | | Human | PMID:17284590 |
| Biological Process | GO:0032922 | circadian regulation of gene expression | Rbm4 | IMP | | Mouse | PMID:17264215 |
| Biological Process | GO:0097167 | circadian regulation of translation | Rbm4 | IDA | | Mouse | MGI:MGI:3703208|PMID:17264215 |
| Biological Process | GO:0035883 | enteroendocrine cell differentiation | Rbm4 | IMP | MGI:MGI:5467987 | Mouse | PMID:23129807 |
| Biological Process | GO:0035883 | enteroendocrine cell differentiation | Rbm4 | IMP | MGI:MGI:5467988 | Mouse | PMID:23129807 |
| Biological Process | GO:0043153 | entrainment of circadian clock by photoperiod | Rbm4 | IDA | | Mouse | MGI:MGI:3703208|PMID:17264215 |
| Biological Process | GO:0042593 | glucose homeostasis | Rbm4 | IMP | MGI:MGI:5467987 | Mouse | PMID:23129807 |
| Biological Process | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | Rbm4 | IMP | MGI:MGI:5467988 | Mouse | PMID:23129807 |
| Biological Process | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | Rbm4 | IMP | MGI:MGI:5467987 | Mouse | PMID:23129807 |
| Biological Process | GO:0002192 | IRES-dependent translational initiation of linear mRNA | RBM4 | IDA | | Human | PMID:17284590 |
| Biological Process | GO:0035278 | miRNA-mediated gene silencing by inhibition of translation | RBM4 | IDA | | Human | PMID:19801630 |
| Biological Process | GO:0017148 | negative regulation of translation | RBM4 | IDA | | Human | PMID:17284590 |
| Biological Process | GO:0032055 | negative regulation of translation in response to stress | RBM4 | IDA | | Human | PMID:17284590 |
| Biological Process | GO:0045947 | negative regulation of translational initiation | RBM4 | IDA | | Human | PMID:19801630 |
| Biological Process | GO:0031016 | pancreas development | Rbm4 | IMP | MGI:MGI:5467987 | Mouse | PMID:23129807 |
| Biological Process | GO:0031016 | pancreas development | Rbm4 | IMP | MGI:MGI:5467988 | Mouse | PMID:23129807 |
| Biological Process | GO:0051149 | positive regulation of muscle cell differentiation | RBM4 | IDA | | Human | PMID:21518792 |
| Biological Process | GO:0045727 | positive regulation of translation | Rbm4 | IDA | | Mouse | PMID:17264215 |
| Biological Process | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | RBM4 | IDA | | Human | PMID:12628928 |
| Biological Process | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | RBM4 | IDA | | Human | PMID:16260624 |
| Biological Process | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | RBM4 | IDA | | Human | PMID:16777844 |
| Biological Process | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | RBM4 | IDA | | Human | PMID:16934801 |
| Biological Process | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | RBM4 | IDA | | Human | PMID:21518792 |
| Biological Process | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | Rbm4 | IMP | MGI:MGI:5467988 | Mouse | PMID:23129807 |
| Biological Process | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | Rbm4 | IMP | MGI:MGI:5467987 | Mouse | PMID:23129807 |
| Biological Process | GO:0010468 | regulation of gene expression | Rbm4 | IMP | MGI:MGI:5467988 | Mouse | PMID:23129807 |
| Biological Process | GO:0010468 | regulation of gene expression | Rbm4 | IMP | MGI:MGI:5467987 | Mouse | PMID:23129807 |
| Biological Process | GO:0046626 | regulation of insulin receptor signaling pathway | Rbm4 | IDA | | Mouse | PMID:23129807 |
| Biological Process | GO:0046822 | regulation of nucleocytoplasmic transport | RBM4 | IDA | | Human | PMID:12628928 |
| Biological Process | GO:0046822 | regulation of nucleocytoplasmic transport | RBM4 | IDA | | Human | PMID:17284590 |
| Biological Process | GO:0046822 | regulation of nucleocytoplasmic transport | Rbm4 | IDA | | Mouse | MGI:MGI:4436983|PMID:19801630 |
| Biological Process | GO:0046685 | response to arsenic-containing substance | RBM4 | IDA | | Human | PMID:17284590 |
 Analysis Tools
Analysis Tools