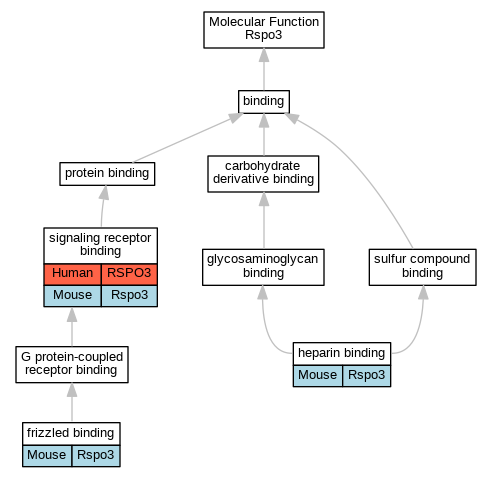
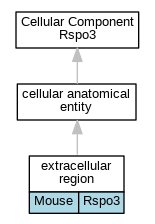
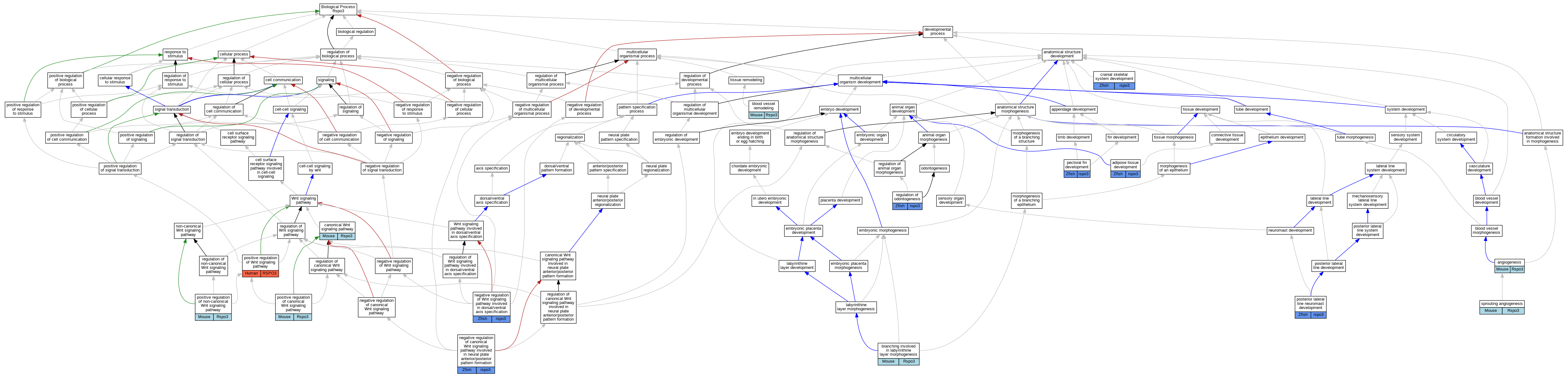
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005109 | frizzled binding | Rspo3 | IPI | PR:Q61091 | Mouse | PMID:16543246 |
| Molecular Function | GO:0008201 | heparin binding | Rspo3 | IDA | | Mouse | PMID:16543246 |
| Molecular Function | GO:0005102 | signaling receptor binding | RSPO3 | IPI | UniProtKB:Q9HBX8 | Human | PMID:22615920 |
| Molecular Function | GO:0005102 | signaling receptor binding | Rspo3 | IPI | UniProtKB:O75581 | Mouse | PMID:16543246 |
| Cellular Component | GO:0005576 | extracellular region | Rspo3 | IDA | | Mouse | PMID:16543246 |
| Biological Process | GO:0060612 | adipose tissue development | rspo3 | IMP | ZFIN:ZDB-GENO-210819-2 | Zfish | PMID:32493999 |
| Biological Process | GO:0001525 | angiogenesis | Rspo3 | IMP | MGI:MGI:5609537 | Mouse | PMID:18842812 |
| Biological Process | GO:0001974 | blood vessel remodeling | Rspo3 | IMP | | Mouse | MGI:MGI:5751613|PMID:26766444 |
| Biological Process | GO:0060670 | branching involved in labyrinthine layer morphogenesis | Rspo3 | IMP | MGI:MGI:3695687 | Mouse | PMID:16963017 |
| Biological Process | GO:0060070 | canonical Wnt signaling pathway | Rspo3 | IMP | MGI:MGI:5609537 | Mouse | PMID:18842812 |
| Biological Process | GO:1904888 | cranial skeletal system development | rspo3 | IGI | ZFIN:ZDB-GENO-221013-7,ZFIN:ZDB-CRISPR-211118-1,ZFIN:ZDB-CRISPR-211118-2,ZFIN:ZDB-CRISPR-211118-3,ZFIN:ZDB-CRISPR-211118-4 | Zfish | PMID:33712657 |
| Biological Process | GO:1904888 | cranial skeletal system development | rspo3 | IGI | ZFIN:ZDB-GENO-221013-6,ZFIN:ZDB-CRISPR-211118-1,ZFIN:ZDB-CRISPR-211118-2,ZFIN:ZDB-CRISPR-211118-3,ZFIN:ZDB-CRISPR-211118-4 | Zfish | PMID:33712657 |
| Biological Process | GO:1904888 | cranial skeletal system development | rspo3 | IMP | ZFIN:ZDB-GENO-221013-6 | Zfish | PMID:33712657 |
| Biological Process | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation | rspo3 | IMP | ZFIN:ZDB-MRPHLNO-140929-7 | Zfish | PMID:24918770 |
| Biological Process | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation | rspo3 | IMP | ZFIN:ZDB-MRPHLNO-140929-8 | Zfish | PMID:24918770 |
| Biological Process | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification | rspo3 | IMP | ZFIN:ZDB-MRPHLNO-140929-8 | Zfish | PMID:24918770 |
| Biological Process | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification | rspo3 | IMP | ZFIN:ZDB-MRPHLNO-140929-7 | Zfish | PMID:24918770 |
| Biological Process | GO:0033339 | pectoral fin development | rspo3 | IGI | ZFIN:ZDB-GENO-221013-7,ZFIN:ZDB-CRISPR-211118-1,ZFIN:ZDB-CRISPR-211118-2,ZFIN:ZDB-CRISPR-211118-3,ZFIN:ZDB-CRISPR-211118-4 | Zfish | PMID:33712657 |
| Biological Process | GO:0033339 | pectoral fin development | rspo3 | IGI | ZFIN:ZDB-GENO-221013-6,ZFIN:ZDB-CRISPR-211118-1,ZFIN:ZDB-CRISPR-211118-2,ZFIN:ZDB-CRISPR-211118-3,ZFIN:ZDB-CRISPR-211118-4 | Zfish | PMID:33712657 |
| Biological Process | GO:0090263 | positive regulation of canonical Wnt signaling pathway | Rspo3 | IMP | MGI:MGI:5609537 | Mouse | PMID:18842812 |
| Biological Process | GO:0090263 | positive regulation of canonical Wnt signaling pathway | Rspo3 | IDA | | Mouse | PMID:16543246 |
| Biological Process | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway | Rspo3 | IMP | | Mouse | MGI:MGI:5751613|PMID:26766444 |
| Biological Process | GO:0030177 | positive regulation of Wnt signaling pathway | RSPO3 | IDA | | Human | PMID:29769720 |
| Biological Process | GO:0048919 | posterior lateral line neuromast development | rspo3 | IMP | ZFIN:ZDB-MRPHLNO-150114-1 | Zfish | PMID:23637337 |
| Biological Process | GO:0042481 | regulation of odontogenesis | rspo3 | IGI | ZFIN:ZDB-GENO-221013-7,ZFIN:ZDB-CRISPR-211118-1,ZFIN:ZDB-CRISPR-211118-2,ZFIN:ZDB-CRISPR-211118-3,ZFIN:ZDB-CRISPR-211118-4 | Zfish | PMID:33712657 |
| Biological Process | GO:0042481 | regulation of odontogenesis | rspo3 | IGI | ZFIN:ZDB-GENO-221013-6,ZFIN:ZDB-CRISPR-211118-1,ZFIN:ZDB-CRISPR-211118-2,ZFIN:ZDB-CRISPR-211118-3,ZFIN:ZDB-CRISPR-211118-4 | Zfish | PMID:33712657 |
| Biological Process | GO:0042481 | regulation of odontogenesis | rspo3 | IMP | ZFIN:ZDB-GENO-221013-6 | Zfish | PMID:33712657 |
| Biological Process | GO:0002040 | sprouting angiogenesis | Rspo3 | IMP | | Mouse | MGI:MGI:5751613|PMID:26766444 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools