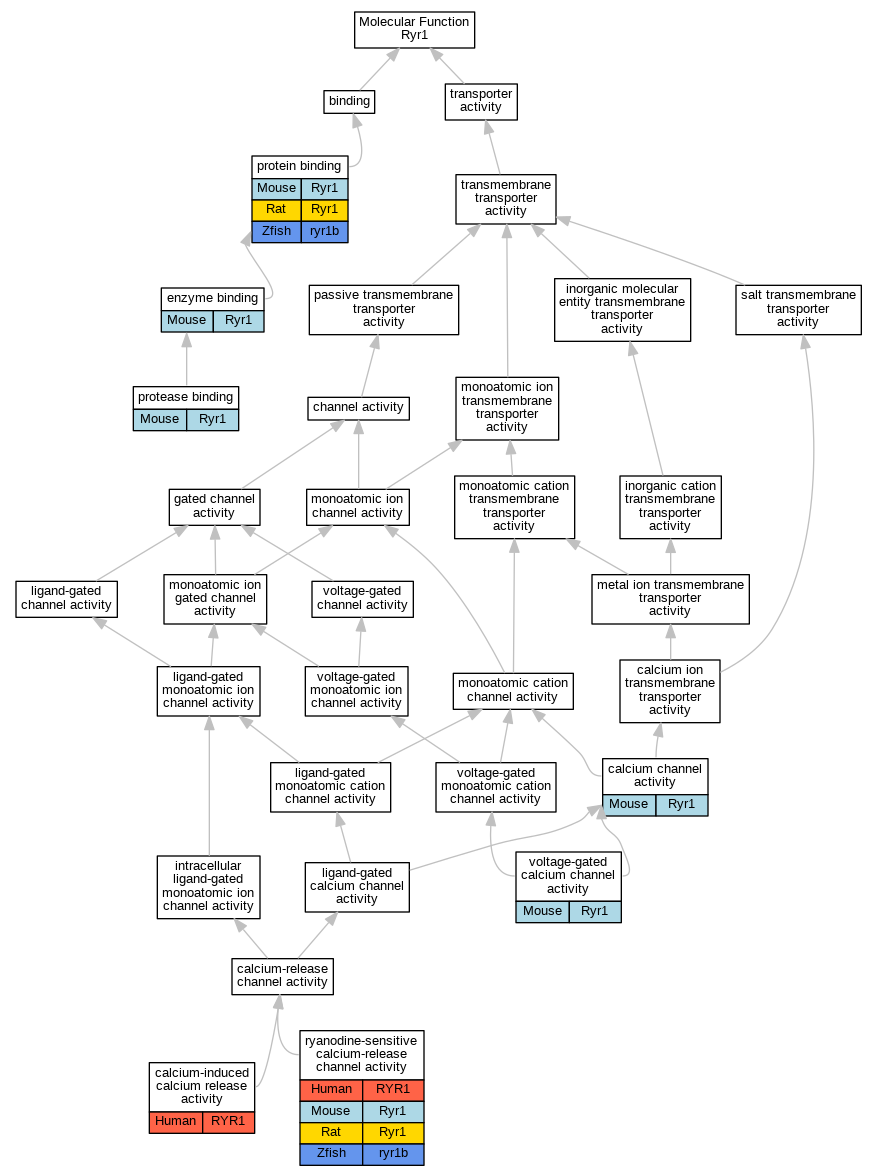
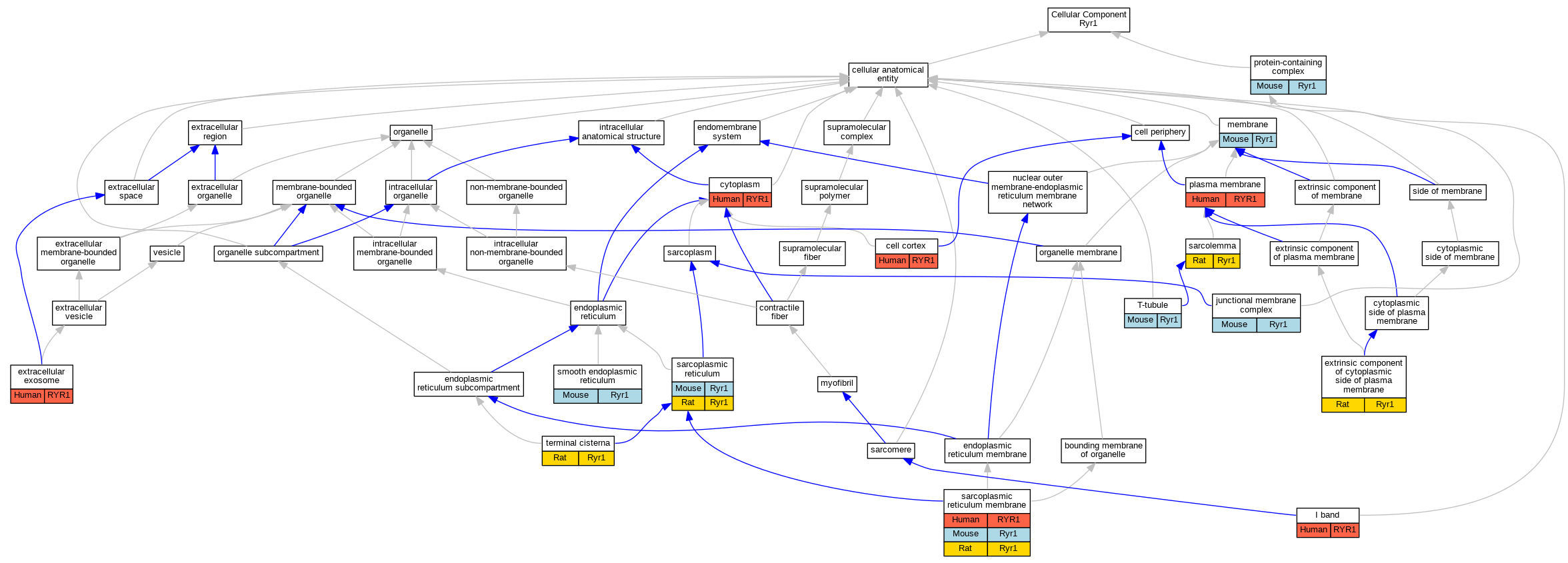
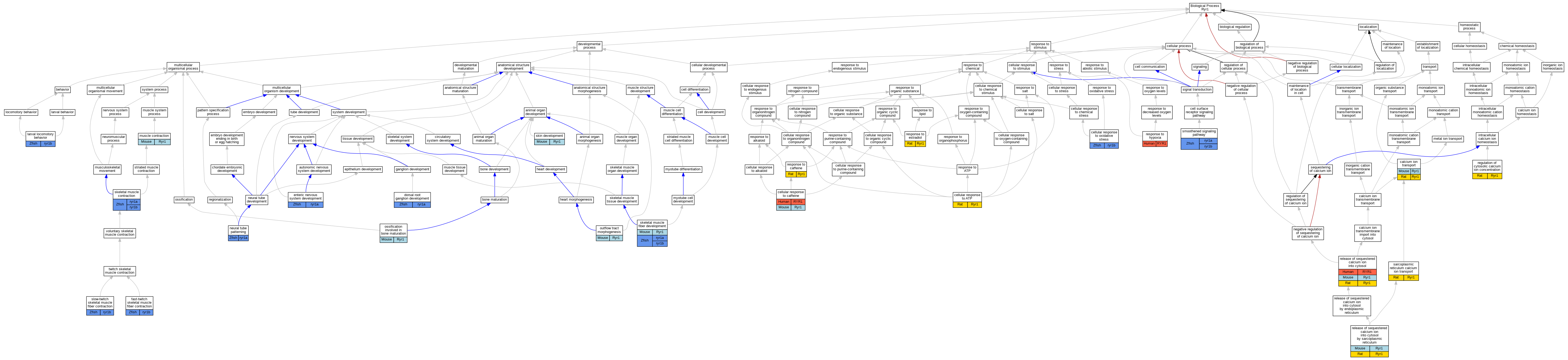
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005262 | calcium channel activity | Ryr1 | IMP | | Mouse | MGI:MGI:3764002|PMID:18003898 |
| Molecular Function | GO:0048763 | calcium-induced calcium release activity | RYR1 | IMP | | Human | PMID:26115329 |
| Molecular Function | GO:0019899 | enzyme binding | Ryr1 | IPI | UniProtKB:P26883 | Mouse | MGI:MGI:3837615|PMID:19198614 |
| Molecular Function | GO:0002020 | protease binding | Ryr1 | IPI | UniProtKB:P16259 | Mouse | MGI:MGI:3811664|PMID:18676612 |
| Molecular Function | GO:0005515 | protein binding | Ryr1 | IPI | UniProtKB:P26883 | Mouse | MGI:MGI:3775502|PMID:18268335 |
| Molecular Function | GO:0005515 | protein binding | Ryr1 | IPI | UniProtKB:O55108|UniProtKB:P49070 | Mouse | MGI:MGI:3688344|PMID:15102471 |
| Molecular Function | GO:0005515 | protein binding | Ryr1 | IPI | UniProtKB:P26883 | Mouse | MGI:MGI:5438859|PMID:22939628 |
| Molecular Function | GO:0005515 | protein binding | Ryr1 | IPI | RGD:619856 | Rat | PMID:16176928|RGD:7327230 |
| Molecular Function | GO:0005515 | protein binding | ryr1b | IPI | ZFIN:ZDB-GENE-040801-248 | Zfish | PMID:23736855 |
| Molecular Function | GO:0005219 | ryanodine-sensitive calcium-release channel activity | RYR1 | IMP | | Human | PMID:11741831 |
| Molecular Function | GO:0005219 | ryanodine-sensitive calcium-release channel activity | RYR1 | IMP | | Human | PMID:16163667 |
| Molecular Function | GO:0005219 | ryanodine-sensitive calcium-release channel activity | RYR1 | IDA | | Human | PMID:22480578 |
| Molecular Function | GO:0005219 | ryanodine-sensitive calcium-release channel activity | RYR1 | IMP | | Human | PMID:26115329 |
| Molecular Function | GO:0005219 | ryanodine-sensitive calcium-release channel activity | Ryr1 | IMP | | Mouse | MGI:MGI:74313|PMID:7621815 |
| Molecular Function | GO:0005219 | ryanodine-sensitive calcium-release channel activity | Ryr1 | IDA | | Rat | PMID:11316255|RGD:12903237 |
| Molecular Function | GO:0005219 | ryanodine-sensitive calcium-release channel activity | Ryr1 | IDA | | Rat | PMID:19116207|RGD:7242196 |
| Molecular Function | GO:0005219 | ryanodine-sensitive calcium-release channel activity | Ryr1 | IDA | | Rat | PMID:19230144|RGD:7175256 |
| Molecular Function | GO:0005219 | ryanodine-sensitive calcium-release channel activity | ryr1b | IMP | ZFIN:ZDB-GENO-070928-2 | Zfish | PMID:17596281 |
| Molecular Function | GO:0005245 | voltage-gated calcium channel activity | Ryr1 | IDA | | Mouse | MGI:MGI:3764002|PMID:18003898 |
| Molecular Function | GO:0005245 | voltage-gated calcium channel activity | Ryr1 | IDA | | Mouse | PMID:12954602 |
| Cellular Component | GO:0005938 | cell cortex | RYR1 | IDA | | Human | PMID:11206130 |
| Cellular Component | GO:0005737 | cytoplasm | RYR1 | IDA | | Human | PMID:11206130 |
| Cellular Component | GO:0070062 | extracellular exosome | RYR1 | HDA | | Human | PMID:19056867 |
| Cellular Component | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane | Ryr1 | IDA | | Rat | PMID:20177054|RGD:7242274 |
| Cellular Component | GO:0031674 | I band | RYR1 | IDA | | Human | PMID:11206130 |
| Cellular Component | GO:0030314 | junctional membrane complex | Ryr1 | IDA | | Mouse | PMID:10444070 |
| Cellular Component | GO:0016020 | membrane | Ryr1 | IDA | | Mouse | PMID:12704193 |
| Cellular Component | GO:0016020 | membrane | Ryr1 | IDA | | Mouse | PMID:7832748 |
| Cellular Component | GO:0005886 | plasma membrane | RYR1 | IDA | | Human | PMID:11206130 |
| Cellular Component | GO:0032991 | protein-containing complex | Ryr1 | IDA | | Mouse | MGI:MGI:3811664|PMID:18676612 |
| Cellular Component | GO:0042383 | sarcolemma | Ryr1 | IDA | | Rat | PMID:21474431|RGD:7175530 |
| Cellular Component | GO:0016529 | sarcoplasmic reticulum | Ryr1 | IDA | | Mouse | PMID:8943043 |
| Cellular Component | GO:0016529 | sarcoplasmic reticulum | Ryr1 | IDA | | Mouse | PMID:7832748 |
| Cellular Component | GO:0016529 | sarcoplasmic reticulum | Ryr1 | IDA | | Mouse | PMID:15536090 |
| Cellular Component | GO:0016529 | sarcoplasmic reticulum | Ryr1 | IDA | | Mouse | PMID:11784029 |
| Cellular Component | GO:0016529 | sarcoplasmic reticulum | Ryr1 | IDA | | Mouse | PMID:12954602 |
| Cellular Component | GO:0016529 | sarcoplasmic reticulum | Ryr1 | IDA | | Rat | PMID:19230144|RGD:7175256 |
| Cellular Component | GO:0033017 | sarcoplasmic reticulum membrane | RYR1 | IDA | | Human | PMID:7556644 |
| Cellular Component | GO:0033017 | sarcoplasmic reticulum membrane | Ryr1 | IDA | | Mouse | MGI:MGI:3764002|PMID:18003898 |
| Cellular Component | GO:0033017 | sarcoplasmic reticulum membrane | Ryr1 | IDA | | Mouse | PMID:14592808 |
| Cellular Component | GO:0033017 | sarcoplasmic reticulum membrane | Ryr1 | IDA | | Rat | PMID:11316255|RGD:12903237 |
| Cellular Component | GO:0033017 | sarcoplasmic reticulum membrane | Ryr1 | IDA | | Rat | PMID:21454501|RGD:8554079 |
| Cellular Component | GO:0005790 | smooth endoplasmic reticulum | Ryr1 | IDA | | Mouse | PMID:23454728 |
| Cellular Component | GO:0030315 | T-tubule | Ryr1 | IDA | | Mouse | PMID:23695157 |
| Cellular Component | GO:0030315 | T-tubule | Ryr1 | IDA | | Mouse | PMID:15536090 |
| Cellular Component | GO:0030315 | T-tubule | Ryr1 | IDA | | Mouse | PMID:8943043 |
| Cellular Component | GO:0030315 | T-tubule | Ryr1 | IDA | | Mouse | PMID:22355118 |
| Cellular Component | GO:0014802 | terminal cisterna | Ryr1 | IDA | | Rat | PMID:11316255|RGD:12903237 |
| Biological Process | GO:0006816 | calcium ion transport | Ryr1 | IDA | | Mouse | PMID:12954602 |
| Biological Process | GO:0006816 | calcium ion transport | Ryr1 | IDA | | Rat | PMID:19116207|RGD:7242196 |
| Biological Process | GO:0071318 | cellular response to ATP | Ryr1 | IDA | | Rat | PMID:11316255|RGD:12903237 |
| Biological Process | GO:0071313 | cellular response to caffeine | RYR1 | IMP | | Human | PMID:16163667 |
| Biological Process | GO:0071313 | cellular response to caffeine | RYR1 | IMP | | Human | PMID:26115329 |
| Biological Process | GO:0071313 | cellular response to caffeine | Ryr1 | IMP | | Mouse | MGI:MGI:74313|PMID:7621815 |
| Biological Process | GO:0034599 | cellular response to oxidative stress | ryr1b | IMP | | Zfish | PMID:22418739 |
| Biological Process | GO:1990791 | dorsal root ganglion development | ryr1a | IMP | ZFIN:ZDB-MRPHLNO-090108-5,ZFIN:ZDB-MRPHLNO-090109-2 | Zfish | PMID:29754802 |
| Biological Process | GO:1990791 | dorsal root ganglion development | ryr1a | IGI | ZFIN:ZDB-GENE-041001-165 | Zfish | PMID:29754802 |
| Biological Process | GO:0048484 | enteric nervous system development | ryr1a | IMP | ZFIN:ZDB-MRPHLNO-090108-5,ZFIN:ZDB-MRPHLNO-090109-2 | Zfish | PMID:29754802 |
| Biological Process | GO:0048484 | enteric nervous system development | ryr1a | IGI | ZFIN:ZDB-GENE-041001-165 | Zfish | PMID:29754802 |
| Biological Process | GO:0031443 | fast-twitch skeletal muscle fiber contraction | ryr1b | IMP | ZFIN:ZDB-MRPHLNO-110822-2 | Zfish | PMID:22075003 |
| Biological Process | GO:0031443 | fast-twitch skeletal muscle fiber contraction | ryr1b | IMP | ZFIN:ZDB-GENO-200723-2 | Zfish | PMID:31383689 |
| Biological Process | GO:0031443 | fast-twitch skeletal muscle fiber contraction | ryr1b | IMP | ZFIN:ZDB-GENO-070928-2 | Zfish | PMID:17596281 |
| Biological Process | GO:0008345 | larval locomotory behavior | ryr1b | IMP | | Zfish | PMID:22418739 |
| Biological Process | GO:0006936 | muscle contraction | Ryr1 | IMP | | Mouse | MGI:MGI:67115|PMID:7515481 |
| Biological Process | GO:0006936 | muscle contraction | Ryr1 | IDA | | Mouse | PMID:12954602 |
| Biological Process | GO:0006936 | muscle contraction | Ryr1 | IMP | MGI:MGI:2152826 | Mouse | PMID:12704193 |
| Biological Process | GO:0021532 | neural tube patterning | ryr1a | IGI | ZFIN:ZDB-GENE-071001-1,ZFIN:ZDB-GENE-041001-165 | Zfish | PMID:29754802 |
| Biological Process | GO:0021532 | neural tube patterning | ryr1a | IMP | ZFIN:ZDB-GENO-190227-9 | Zfish | PMID:29754802 |
| Biological Process | GO:0043931 | ossification involved in bone maturation | Ryr1 | IMP | | Mouse | MGI:MGI:3764002|PMID:18003898 |
| Biological Process | GO:0003151 | outflow tract morphogenesis | Ryr1 | IMP | | Mouse | MGI:MGI:3764002|PMID:18003898 |
| Biological Process | GO:0051480 | regulation of cytosolic calcium ion concentration | Ryr1 | IDA | | Rat | PMID:19116207|RGD:7242196 |
| Biological Process | GO:0051209 | release of sequestered calcium ion into cytosol | RYR1 | IMP | | Human | PMID:11741831 |
| Biological Process | GO:0051209 | release of sequestered calcium ion into cytosol | RYR1 | IMP | | Human | PMID:16163667 |
| Biological Process | GO:0051209 | release of sequestered calcium ion into cytosol | Ryr1 | IMP | MGI:MGI:2152826 | Mouse | PMID:12704193 |
| Biological Process | GO:0051209 | release of sequestered calcium ion into cytosol | Ryr1 | IDA | | Rat | PMID:19116207|RGD:7242196 |
| Biological Process | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | Ryr1 | IMP | | Mouse | MGI:MGI:3764002|PMID:18003898 |
| Biological Process | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | Ryr1 | IDA | | Rat | PMID:11316255|RGD:12903237 |
| Biological Process | GO:0031000 | response to caffeine | Ryr1 | IDA | | Rat | PMID:19116207|RGD:7242196 |
| Biological Process | GO:0032355 | response to estradiol | Ryr1 | IEP | | Rat | PMID:19593990|RGD:13782367 |
| Biological Process | GO:0001666 | response to hypoxia | RYR1 | IDA | | Human | PMID:19120137 |
| Biological Process | GO:0070296 | sarcoplasmic reticulum calcium ion transport | Ryr1 | IDA | | Rat | PMID:19230144|RGD:7175256 |
| Biological Process | GO:0003009 | skeletal muscle contraction | ryr1a | IMP | ZFIN:ZDB-GENO-200729-3 | Zfish | PMID:31383689 |
| Biological Process | GO:0003009 | skeletal muscle contraction | ryr1b | IMP | ZFIN:ZDB-GENO-200729-3 | Zfish | PMID:31383689 |
| Biological Process | GO:0048741 | skeletal muscle fiber development | Ryr1 | IMP | | Mouse | MGI:MGI:3764002|PMID:18003898 |
| Biological Process | GO:0048741 | skeletal muscle fiber development | ryr1a | IMP | ZFIN:ZDB-GENO-190227-2 | Zfish | PMID:29754802 |
| Biological Process | GO:0048741 | skeletal muscle fiber development | ryr1a | IGI | ZFIN:ZDB-GENE-070705-417,ZFIN:ZDB-GENE-041001-165 | Zfish | PMID:29754802 |
| Biological Process | GO:0048741 | skeletal muscle fiber development | ryr1b | IMP | ZFIN:ZDB-MRPHLNO-090108-5,ZFIN:ZDB-MRPHLNO-090109-2 | Zfish | PMID:29754802 |
| Biological Process | GO:0048741 | skeletal muscle fiber development | ryr1b | IMP | ZFIN:ZDB-GENO-190227-2 | Zfish | PMID:29754802 |
| Biological Process | GO:0048741 | skeletal muscle fiber development | ryr1a | IMP | ZFIN:ZDB-MRPHLNO-090108-5,ZFIN:ZDB-MRPHLNO-090109-2 | Zfish | PMID:29754802 |
| Biological Process | GO:0048741 | skeletal muscle fiber development | ryr1b | IGI | ZFIN:ZDB-GENE-020108-2,ZFIN:ZDB-GENE-041001-165 | Zfish | PMID:29754802 |
| Biological Process | GO:0043588 | skin development | Ryr1 | IMP | | Mouse | MGI:MGI:3764002|PMID:18003898 |
| Biological Process | GO:0031444 | slow-twitch skeletal muscle fiber contraction | ryr1b | IMP | ZFIN:ZDB-GENO-200723-2 | Zfish | PMID:31383689 |
| Biological Process | GO:0007224 | smoothened signaling pathway | ryr1a | IGI | ZFIN:ZDB-GENE-071001-1,ZFIN:ZDB-GENE-041001-165 | Zfish | PMID:29754802 |
| Biological Process | GO:0007224 | smoothened signaling pathway | ryr1a | IMP | ZFIN:ZDB-MRPHLNO-090108-5,ZFIN:ZDB-MRPHLNO-090109-2 | Zfish | PMID:29754802 |
| Biological Process | GO:0007224 | smoothened signaling pathway | ryr1b | IGI | ZFIN:ZDB-GENE-020108-2,ZFIN:ZDB-GENE-041001-165 | Zfish | PMID:29754802 |
| Biological Process | GO:0007224 | smoothened signaling pathway | ryr1a | IGI | ZFIN:ZDB-GENE-041001-165 | Zfish | PMID:29754802 |
| Biological Process | GO:0007224 | smoothened signaling pathway | ryr1a | IMP | ZFIN:ZDB-GENO-190227-9 | Zfish | PMID:29754802 |
| Biological Process | GO:0007224 | smoothened signaling pathway | ryr1a | IMP | ZFIN:ZDB-GENO-190227-2 | Zfish | PMID:29754802 |
| Biological Process | GO:0007224 | smoothened signaling pathway | ryr1a | IGI | ZFIN:ZDB-GENE-070705-417,ZFIN:ZDB-GENE-041001-165 | Zfish | PMID:29754802 |
| Biological Process | GO:0007224 | smoothened signaling pathway | ryr1b | IMP | ZFIN:ZDB-GENO-190227-2 | Zfish | PMID:29754802 |
 Analysis Tools
Analysis Tools