| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
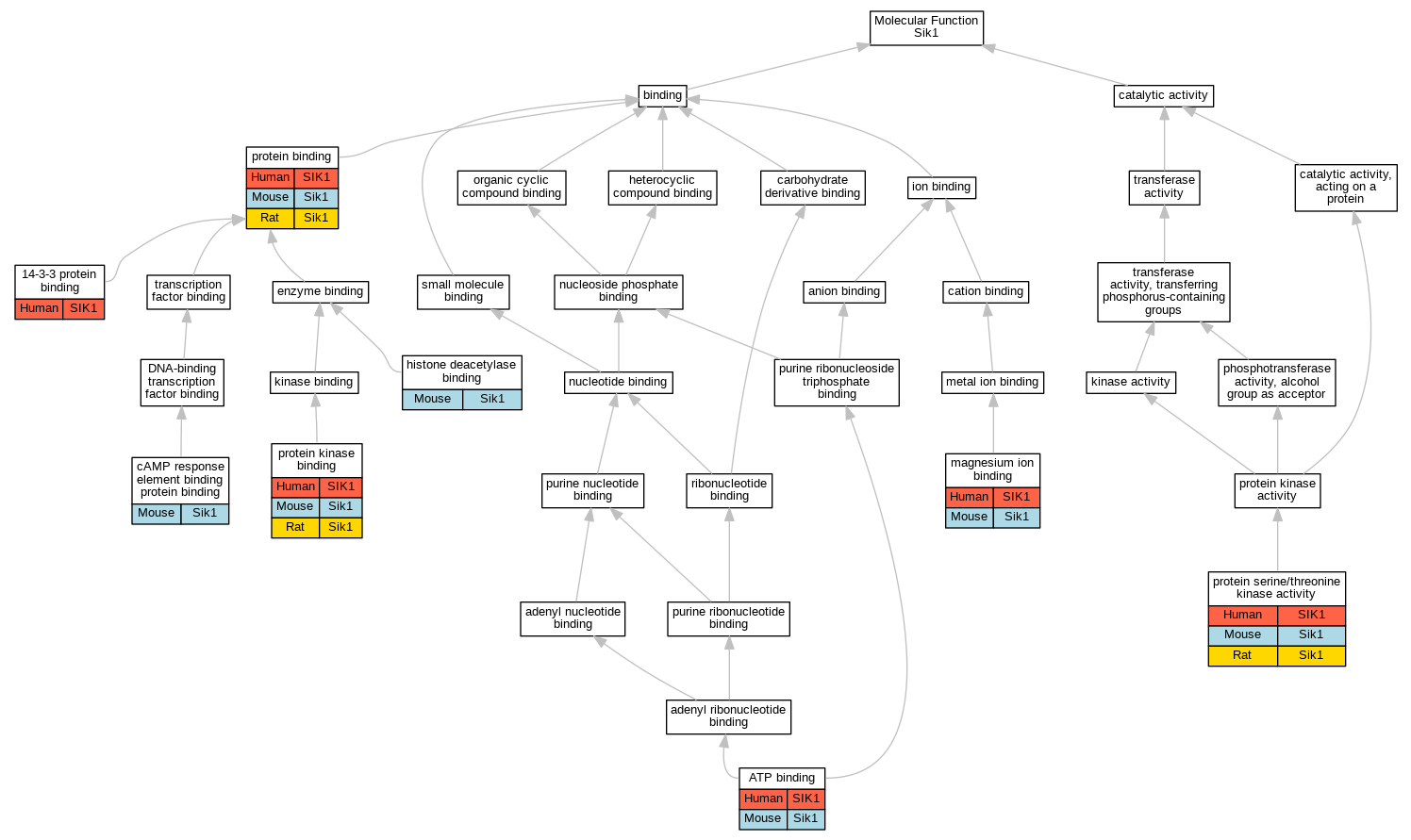
| Molecular Function | GO:0071889 | 14-3-3 protein binding | SIK1 | IDA | | Human | PMID:18348280 |
| Molecular Function | GO:0005524 | ATP binding | SIK1 | IDA | | Human | PMID:14976552 |
| Molecular Function | GO:0005524 | ATP binding | Sik1 | IDA | | Mouse | MGI:MGI:3044497|PMID:15177563 |
| Molecular Function | GO:0008140 | cAMP response element binding protein binding | Sik1 | IDA | | Mouse | MGI:MGI:2389532|PMID:12200423 |
| Molecular Function | GO:0042826 | histone deacetylase binding | Sik1 | IPI | UniProtKB:Q6NZM9|UniProtKB:Q9Z2V6 | Mouse | MGI:MGI:3712602|PMID:17468767 |
| Molecular Function | GO:0000287 | magnesium ion binding | SIK1 | IDA | | Human | PMID:14976552 |
| Molecular Function | GO:0000287 | magnesium ion binding | Sik1 | IDA | | Mouse | MGI:MGI:3044497|PMID:15177563 |
| Molecular Function | GO:0005515 | protein binding | SIK1 | IPI | UniProtKB:P63104 | Human | PMID:16306228 |
| Molecular Function | GO:0005515 | protein binding | SIK1 | IPI | UniProtKB:P63104 | Human | PMID:32707033 |
| Molecular Function | GO:0005515 | protein binding | Sik1 | IPI | UniProtKB:Q9WTN3 | Mouse | MGI:MGI:5294536|PMID:19244231 |
| Molecular Function | GO:0005515 | protein binding | Sik1 | IPI | UniProtKB:P06685 | Rat | PMID:17939993|RGD:8554378 |
| Molecular Function | GO:0005515 | protein binding | Sik1 | IPI | UniProtKB:Q4FZT2 | Rat | PMID:17939993|RGD:8554378 |
| Molecular Function | GO:0019901 | protein kinase binding | SIK1 | IPI | UniProtKB:P49841 | Human | PMID:18348280 |
| Molecular Function | GO:0019901 | protein kinase binding | Sik1 | IPI | UniProtKB:P05132 | Mouse | MGI:MGI:2389532|PMID:12200423 |
| Molecular Function | GO:0019901 | protein kinase binding | Sik1 | IPI | UniProtKB:Q63450 | Rat | PMID:17939993|RGD:8554378 |
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | SIK1 | IDA | | Human | PMID:14976552 |
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | SIK1 | IDA | | Human | PMID:18348280 |
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | Sik1 | IDA | | Mouse | MGI:MGI:3606826|PMID:16148943 |
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | Sik1 | IDA | | Mouse | MGI:MGI:3044497|PMID:15177563 |
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | Sik1 | IDA | | Mouse | MGI:MGI:3712602|PMID:17468767 |
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | Sik1 | IDA | | Mouse | MGI:MGI:5294536|PMID:19244231 |
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | Sik1 | IDA | | Mouse | PMID:12624099 |
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | Sik1 | IMP | | Rat | PMID:12530631|RGD:1299210 |
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | Sik1 | IDA | | Rat | PMID:17939993|RGD:8554378 |
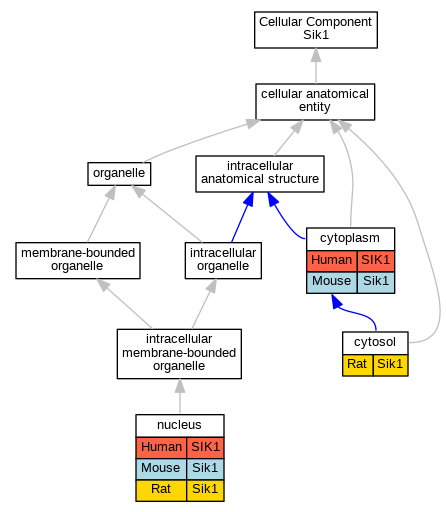
| Cellular Component | GO:0005737 | cytoplasm | SIK1 | IDA | | Human | PMID:16306228 |
| Cellular Component | GO:0005737 | cytoplasm | Sik1 | IDA | | Mouse | MGI:MGI:3710425|PMID:16817901 |
| Cellular Component | GO:0005737 | cytoplasm | Sik1 | IDA | | Mouse | MGI:MGI:2389532|PMID:12200423 |
| Cellular Component | GO:0005737 | cytoplasm | Sik1 | IDA | | Mouse | MGI:MGI:3044497|PMID:15177563 |
| Cellular Component | GO:0005829 | cytosol | Sik1 | IDA | | Rat | PMID:12530631|RGD:1299210 |
| Cellular Component | GO:0005634 | nucleus | SIK1 | IDA | | Human | PMID:16306228 |
| Cellular Component | GO:0005634 | nucleus | Sik1 | IDA | | Mouse | MGI:MGI:3710425|PMID:16817901 |
| Cellular Component | GO:0005634 | nucleus | Sik1 | IDA | | Mouse | MGI:MGI:2389532|PMID:12200423 |
| Cellular Component | GO:0005634 | nucleus | Sik1 | IDA | | Mouse | PMID:12624099 |
| Cellular Component | GO:0005634 | nucleus | Sik1 | IDA | | Rat | PMID:12530631|RGD:1299210 |
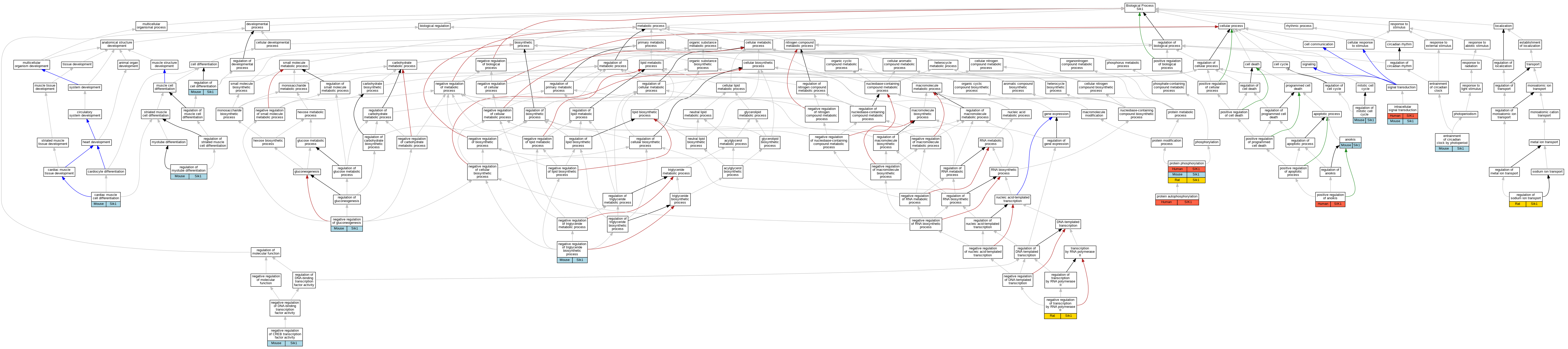
| Biological Process | GO:0043276 | anoikis | Sik1 | IMP | | Mouse | MGI:MGI:4887544|PMID:19622832 |
| Biological Process | GO:0055007 | cardiac muscle cell differentiation | Sik1 | IMP | | Mouse | MGI:MGI:4437512|PMID:20140255 |
| Biological Process | GO:0043153 | entrainment of circadian clock by photoperiod | Sik1 | IMP | | Mouse | MGI:MGI:5532783|PMID:23993098 |
| Biological Process | GO:0035556 | intracellular signal transduction | SIK1 | IDA | | Human | PMID:14976552 |
| Biological Process | GO:0035556 | intracellular signal transduction | Sik1 | IDA | | Mouse | MGI:MGI:3044497|PMID:15177563 |
| Biological Process | GO:0032792 | negative regulation of CREB transcription factor activity | Sik1 | IMP | | Mouse | MGI:MGI:5532783|PMID:23993098 |
| Biological Process | GO:0032792 | negative regulation of CREB transcription factor activity | Sik1 | IDA | | Mouse | MGI:MGI:3606826|PMID:16148943 |
| Biological Process | GO:0032792 | negative regulation of CREB transcription factor activity | Sik1 | IDA | | Mouse | MGI:MGI:2389532|PMID:12200423 |
| Biological Process | GO:0045721 | negative regulation of gluconeogenesis | Sik1 | IDA | | Mouse | MGI:MGI:3606826|PMID:16148943 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | Sik1 | IMP | | Rat | PMID:12530631|RGD:1299210 |
| Biological Process | GO:0010868 | negative regulation of triglyceride biosynthetic process | Sik1 | IDA | | Mouse | MGI:MGI:5294536|PMID:19244231 |
| Biological Process | GO:2000210 | positive regulation of anoikis | SIK1 | IMP | | Human | PMID:19622832 |
| Biological Process | GO:0046777 | protein autophosphorylation | SIK1 | IDA | | Human | PMID:18348280 |
| Biological Process | GO:0006468 | protein phosphorylation | SIK1 | IDA | | Human | PMID:14976552 |
| Biological Process | GO:0006468 | protein phosphorylation | Sik1 | IDA | | Mouse | MGI:MGI:3044497|PMID:15177563 |
| Biological Process | GO:0006468 | protein phosphorylation | Sik1 | IDA | | Mouse | MGI:MGI:5532783|PMID:23993098 |
| Biological Process | GO:0006468 | protein phosphorylation | Sik1 | IDA | | Mouse | PMID:12624099 |
| Biological Process | GO:0006468 | protein phosphorylation | Sik1 | IMP | | Rat | PMID:12530631|RGD:1299210 |
| Biological Process | GO:0045595 | regulation of cell differentiation | Sik1 | IEP | | Mouse | MGI:MGI:3044497|PMID:15177563 |
| Biological Process | GO:0007346 | regulation of mitotic cell cycle | Sik1 | IDA | | Mouse | MGI:MGI:3044497|PMID:15177563 |
| Biological Process | GO:0010830 | regulation of myotube differentiation | Sik1 | IDA | | Mouse | MGI:MGI:3712602|PMID:17468767 |
| Biological Process | GO:0002028 | regulation of sodium ion transport | Sik1 | IDA | | Rat | PMID:17939993|RGD:8554378 |
 Analysis Tools
Analysis Tools