|
Source |
Alliance of Genome Resources |
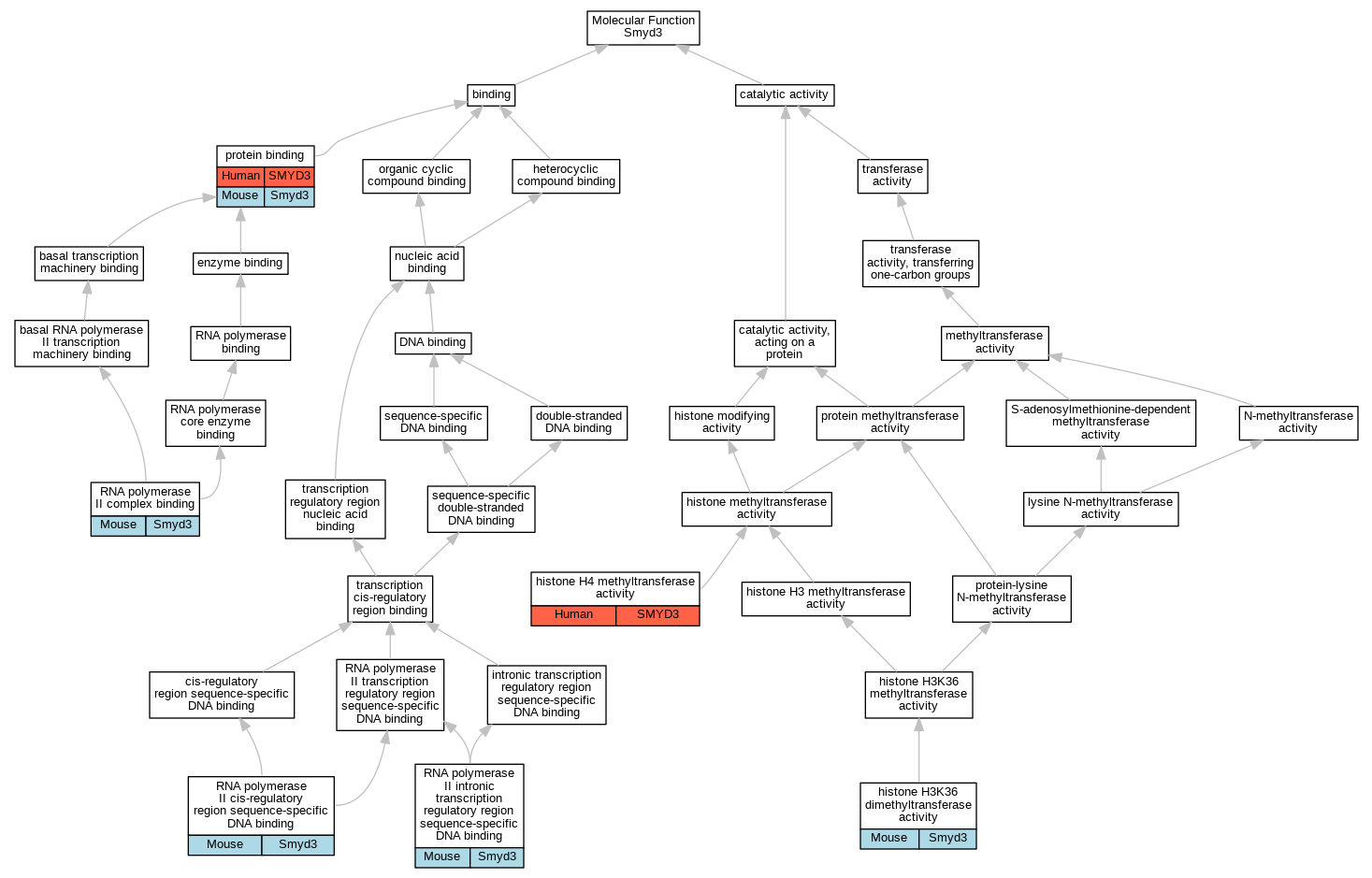
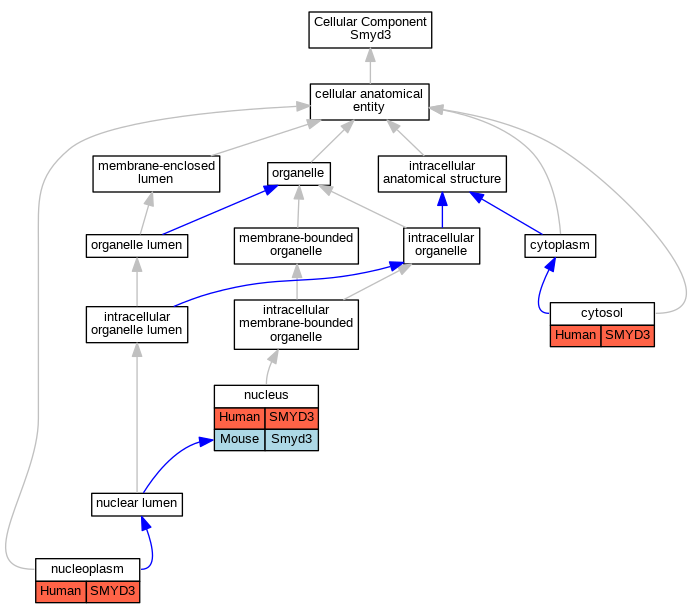
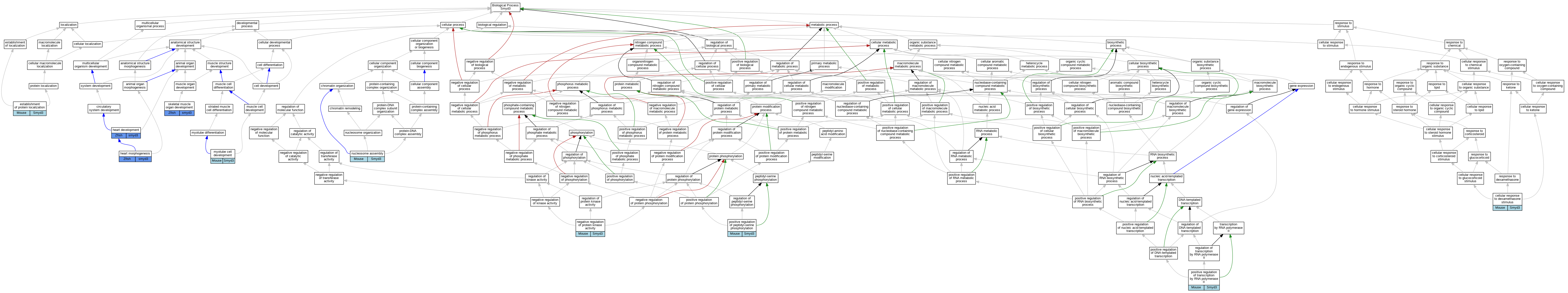
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0140954 | histone H3K36 dimethyltransferase activity | Smyd3 | IDA | Mouse | MGI:MGI:4943402|PMID:16805913 | |
| Molecular Function | GO:0140939 | histone H4 methyltransferase activity | SMYD3 | IMP | Human | PMID:25738358 | |
| Molecular Function | GO:0005515 | protein binding | SMYD3 | IPI | UniProtKB:P07900 | Human | PMID:25738358 |
| Molecular Function | GO:0005515 | protein binding | SMYD3 | IPI | UniProtKB:Q13064 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | SMYD3 | IPI | UniProtKB:Q15915 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | SMYD3 | IPI | UniProtKB:Q16512 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | SMYD3 | IPI | UniProtKB:Q7Z3B4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | SMYD3 | IPI | UniProtKB:Q92529 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | SMYD3 | IPI | UniProtKB:Q9H0I2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | SMYD3 | IPI | UniProtKB:Q9H0L4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | Smyd3 | IPI | UniProtKB:O60885 | Mouse | PMID:23752591 |
| Molecular Function | GO:0005515 | protein binding | Smyd3 | IPI | PR:Q9ESU6 | Mouse | PMID:23752591 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | Smyd3 | IDA | Mouse | PMID:23752591 | |
| Molecular Function | GO:0000993 | RNA polymerase II complex binding | Smyd3 | IDA | Mouse | PMID:23752591 | |
| Molecular Function | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding | Smyd3 | IDA | Mouse | PMID:23752591 | |
| Cellular Component | GO:0005829 | cytosol | SMYD3 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005654 | nucleoplasm | SMYD3 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005634 | nucleus | SMYD3 | IDA | Human | PMID:25738358 | |
| Cellular Component | GO:0005634 | nucleus | Smyd3 | IDA | Mouse | PMID:23752591 | |
| Biological Process | GO:0071549 | cellular response to dexamethasone stimulus | Smyd3 | IDA | Mouse | PMID:23752591 | |
| Biological Process | GO:0045184 | establishment of protein localization | Smyd3 | IMP | Mouse | PMID:23752591 | |
| Biological Process | GO:0007507 | heart development | smyd3 | IMP | ZFIN:ZDB-MRPHLNO-110922-1 | Zfish | PMID:21887258 |
| Biological Process | GO:0007507 | heart development | smyd3 | IMP | ZFIN:ZDB-MRPHLNO-110922-2 | Zfish | PMID:21887258 |
| Biological Process | GO:0003007 | heart morphogenesis | smyd3 | IMP | ZFIN:ZDB-MRPHLNO-151013-5 | Zfish | PMID:25997738 |
| Biological Process | GO:0003007 | heart morphogenesis | smyd3 | IGI | ZFIN:ZDB-MRPHLNO-151013-5,ZFIN:ZDB-MRPHLNO-151013-6 | Zfish | PMID:25997738 |
| Biological Process | GO:0014904 | myotube cell development | Smyd3 | IMP | Mouse | PMID:23752591 | |
| Biological Process | GO:0006469 | negative regulation of protein kinase activity | Smyd3 | IMP | Mouse | PMID:23752591 | |
| Biological Process | GO:0006334 | nucleosome assembly | Smyd3 | IMP | Mouse | PMID:23752591 | |
| Biological Process | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | Smyd3 | IMP | Mouse | PMID:23752591 | |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Smyd3 | IMP | Mouse | PMID:23752591 | |
| Biological Process | GO:0060538 | skeletal muscle organ development | smyd3 | IMP | ZFIN:ZDB-MRPHLNO-110922-1 | Zfish | PMID:21887258 |
| Biological Process | GO:0060538 | skeletal muscle organ development | smyd3 | IMP | ZFIN:ZDB-MRPHLNO-110922-2 | Zfish | PMID:21887258 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 06/12/2024 MGI 6.13 |

|
|
|
||