|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
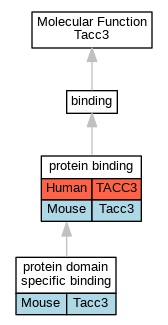
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q14008 | Human | PMID:20360068 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:P09496 | Human | PMID:21297582 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q14008 | Human | PMID:21297582 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:A5D8V6 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q2M2Z5 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q7Z614 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q96Q77 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q9BZE9 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q9NU19 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q14008 | Human | PMID:26496610 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q9NU19 | Human | PMID:29892012 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q00994 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q07866-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q2M2Z5 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q2WGJ6 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q53TS8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q5VTR2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q7Z3B4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q86Y33-5 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q8TC71 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q8TD31-3 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q96DY2-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q96Q77 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q99819 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q99988 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q9BXY8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q9GZM8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | TACC3 | IPI | UniProtKB:Q9NU19 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | Tacc3 | IPI | UniProtKB:O35615 | Mouse | MGI:MGI:3056886|PMID:15234987 |
| Molecular Function | GO:0005515 | protein binding | Tacc3 | IPI | UniProtKB:Q8R1A4 | Mouse | MGI:MGI:5447899|PMID:22842144 |
| Molecular Function | GO:0005515 | protein binding | Tacc3 | IPI | PR:Q9ERR1 | Mouse | PMID:17060449 |
| Molecular Function | GO:0019904 | protein domain specific binding | Tacc3 | IPI | PR:Q7TSG1 | Mouse | PMID:17920017 |
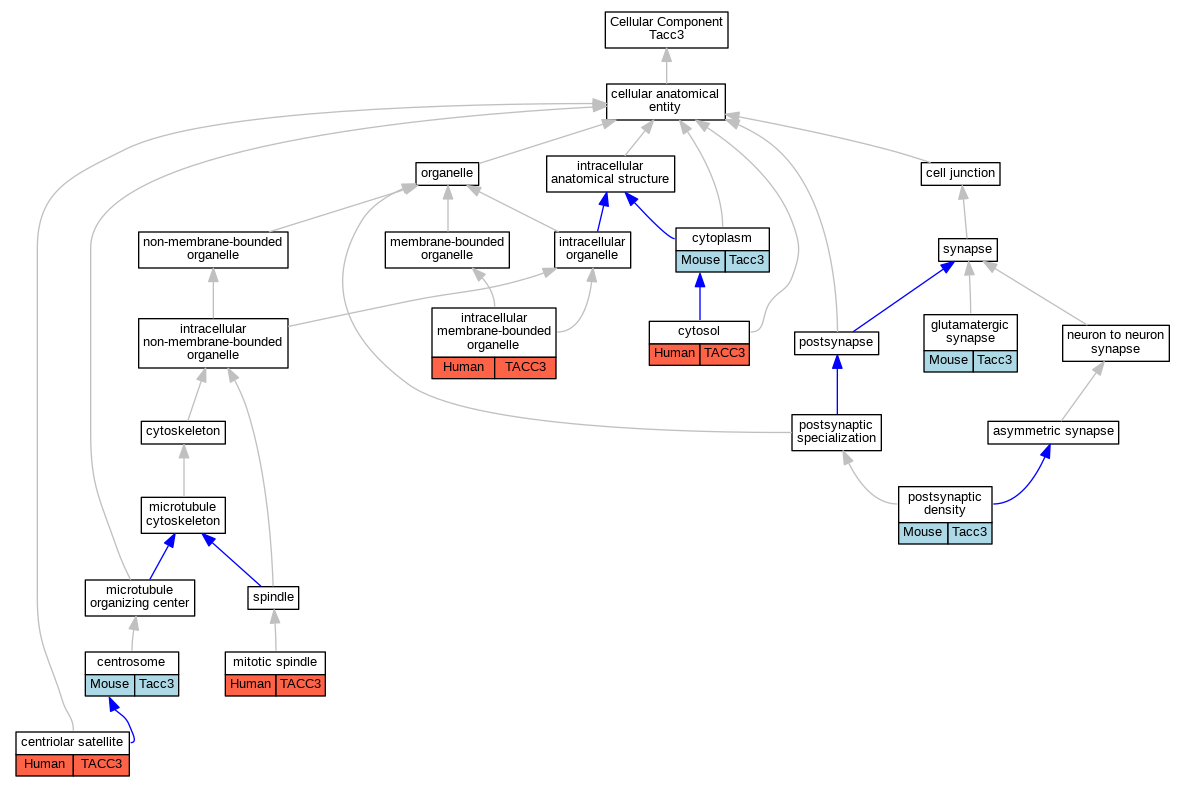
| Cellular Component | GO:0034451 | centriolar satellite | TACC3 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005813 | centrosome | Tacc3 | IDA | Mouse | PMID:17920017 | |
| Cellular Component | GO:0005813 | centrosome | Tacc3 | IDA | Mouse | PMID:11847113 | |
| Cellular Component | GO:0005737 | cytoplasm | Tacc3 | IDA | Mouse | PMID:12237944 | |
| Cellular Component | GO:0005737 | cytoplasm | Tacc3 | IDA | Mouse | PMID:11025203 | |
| Cellular Component | GO:0005829 | cytosol | TACC3 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0098978 | glutamatergic synapse | Tacc3 | IDA | Mouse | PMID:11980711 | |
| Cellular Component | GO:0098978 | glutamatergic synapse | Tacc3 | IEP | Mouse | PMID:11980711 | |
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | TACC3 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0072686 | mitotic spindle | TACC3 | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0014069 | postsynaptic density | Tacc3 | IDA | Mouse | PMID:11980711 | |
| Cellular Component | GO:0014069 | postsynaptic density | Tacc3 | IEP | Mouse | PMID:11980711 | |
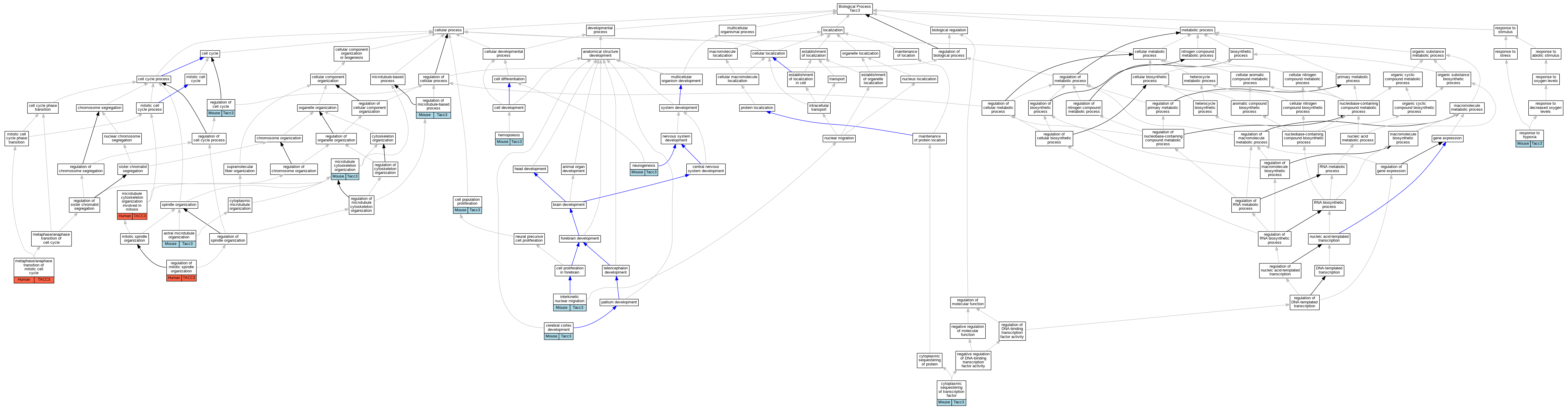
| Biological Process | GO:0030953 | astral microtubule organization | Tacc3 | IDA | Mouse | PMID:17920017 | |
| Biological Process | GO:0008283 | cell population proliferation | Tacc3 | IMP | Mouse | PMID:17920017 | |
| Biological Process | GO:0021987 | cerebral cortex development | Tacc3 | IMP | Mouse | PMID:17920017 | |
| Biological Process | GO:0042994 | cytoplasmic sequestering of transcription factor | Tacc3 | IDA | Mouse | PMID:11025203 | |
| Biological Process | GO:0030097 | hemopoiesis | Tacc3 | IMP | MGI:MGI:2159700 | Mouse | PMID:11847113 |
| Biological Process | GO:0022027 | interkinetic nuclear migration | Tacc3 | IMP | Mouse | PMID:17920017 | |
| Biological Process | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | TACC3 | IMP | Human | PMID:23532825 | |
| Biological Process | GO:0000226 | microtubule cytoskeleton organization | Tacc3 | IMP | Mouse | PMID:17920017 | |
| Biological Process | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | TACC3 | IMP | Human | PMID:23532825 | |
| Biological Process | GO:0022008 | neurogenesis | Tacc3 | IMP | Mouse | PMID:17920017 | |
| Biological Process | GO:0051726 | regulation of cell cycle | Tacc3 | IGI | MGI:MGI:98834 | Mouse | PMID:11847113 |
| Biological Process | GO:0032886 | regulation of microtubule-based process | Tacc3 | IMP | Mouse | PMID:17920017 | |
| Biological Process | GO:0060236 | regulation of mitotic spindle organization | TACC3 | IMP | Human | PMID:21297582 | |
| Biological Process | GO:0001666 | response to hypoxia | Tacc3 | IDA | Mouse | PMID:11025203 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 08/27/2024 MGI 6.24 |

|
|
|
||