Comparative GO Graph (mouse, human, rat, zebrafish)
Compare experimental GO annotations for Human-Mouse-Rat-Zebrafish orthologs to Mouse Thap11
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
- This class contains
- Human genes THAP11
- Mouse genes Thap11
- Rat genes Thap11
- Zfish genes thap11
- Annotations are indicated with nodes colored by organism:
Human,
Mouse,
Rat,
Zfish
- Relations between GO terms are indicated by colored edges "is_a"; "part_of"; "regulates"; "positively_regulates"; "negatively_regulates"
- GO annotations: Human from GOA; Mouse from MGI; Rat from RGD; Zebrafish from ZFIN.
- Only experimental annotations are displayed: EXP, IDA, IEP, IGI, IMP, IPI, HTP, HDA, HMP, HGI, HEP. Evidence codes are listed at the bottom of the page.
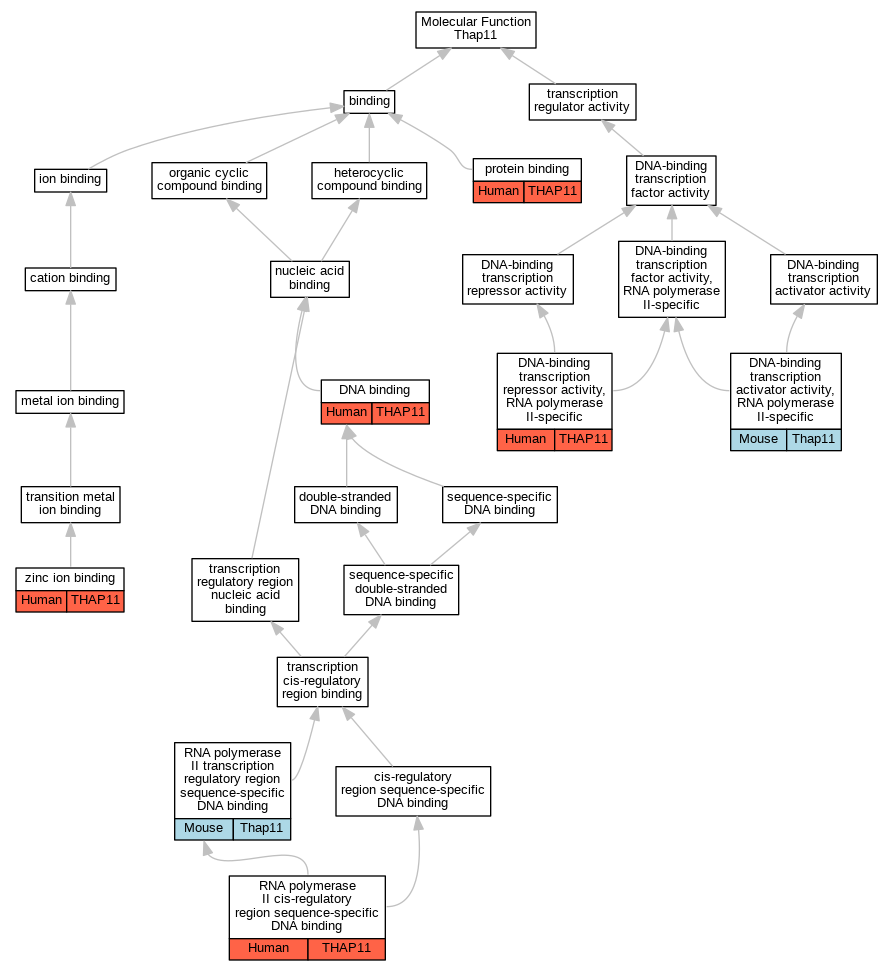
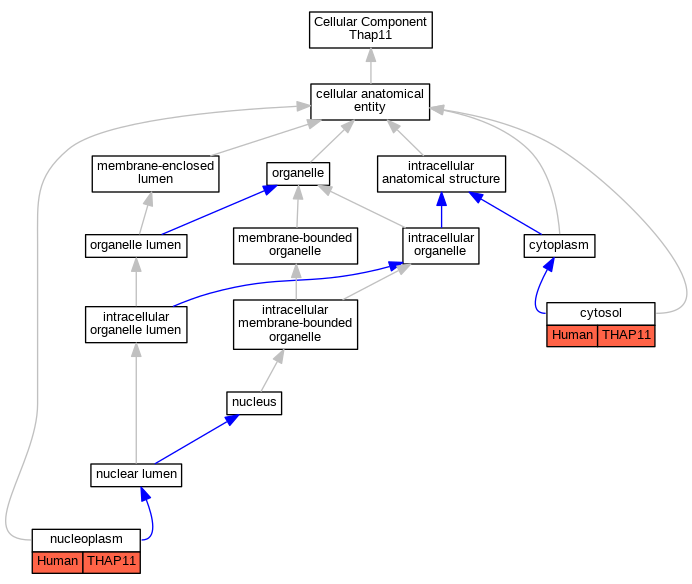
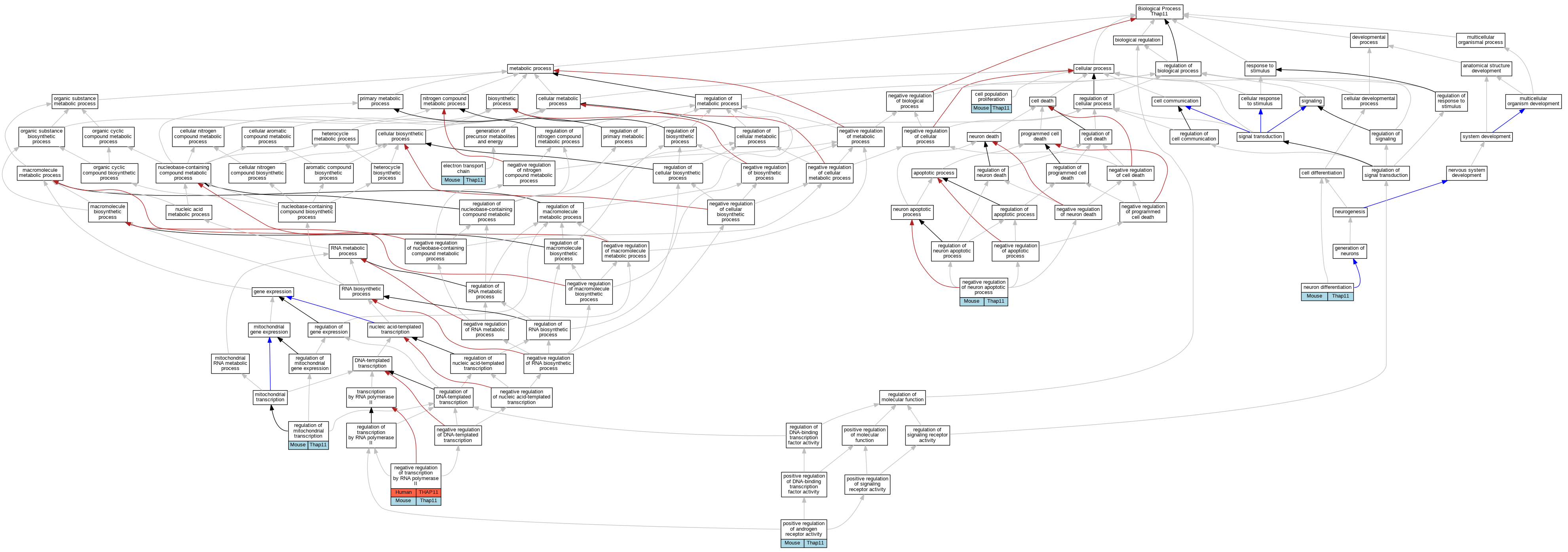 Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Thap11
Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Thap11
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0003677 | DNA binding | THAP11 | IDA | | Human | PMID:23306615 |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | Thap11 | IMP | MGI:MGI:3052237,MGI:MGI:3797582 | Mouse | PMID:26876175 |
| Molecular Function | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | THAP11 | IDA | | Human | PMID:19008924 |
| Molecular Function | GO:0005515 | protein binding | THAP11 | IPI | UniProtKB:P51610 | Human | PMID:18585351 |
| Molecular Function | GO:0005515 | protein binding | THAP11 | IPI | UniProtKB:P70677 | Human | PMID:18585351 |
| Molecular Function | GO:0005515 | protein binding | THAP11 | IPI | UniProtKB:Q13363 | Human | PMID:21988832 |
| Molecular Function | GO:0005515 | protein binding | THAP11 | IPI | UniProtKB:Q15365 | Human | PMID:22673507 |
| Molecular Function | GO:0005515 | protein binding | THAP11 | IPI | UniProtKB:Q8N1B4 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | THAP11 | IPI | UniProtKB:Q13363 | Human | PMID:31041561 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | THAP11 | IDA | | Human | PMID:19008924 |
| Molecular Function | GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | Thap11 | IMP | MGI:MGI:3052237,MGI:MGI:3797582 | Mouse | PMID:26876175 |
| Molecular Function | GO:0008270 | zinc ion binding | THAP11 | IDA | | Human | PMID:23306615 |
| Cellular Component | GO:0005829 | cytosol | THAP11 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005654 | nucleoplasm | THAP11 | IDA | | Human | GO_REF:0000052 |
| Biological Process | GO:0008283 | cell population proliferation | Thap11 | IMP | MGI:MGI:3052237,MGI:MGI:3797582 | Mouse | PMID:26876175 |
| Biological Process | GO:0022900 | electron transport chain | Thap11 | IMP | MGI:MGI:3052237,MGI:MGI:3797582 | Mouse | PMID:26876175 |
| Biological Process | GO:0043524 | negative regulation of neuron apoptotic process | Thap11 | IMP | MGI:MGI:3052237,MGI:MGI:3797582 | Mouse | PMID:26876175 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | THAP11 | IDA | | Human | PMID:19008924 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | Thap11 | IMP | MGI:MGI:3052237,MGI:MGI:3797582 | Mouse | PMID:26876175 |
| Biological Process | GO:0030182 | neuron differentiation | Thap11 | IMP | MGI:MGI:3052237,MGI:MGI:3797582 | Mouse | PMID:26876175 |
| Biological Process | GO:2000825 | positive regulation of androgen receptor activity | Thap11 | IMP | MGI:MGI:3052237,MGI:MGI:3797582 | Mouse | PMID:26876175 |
| Biological Process | GO:1903108 | regulation of mitochondrial transcription | Thap11 | IMP | MGI:MGI:3052237,MGI:MGI:3797582 | Mouse | PMID:26876175 |
Gene Ontology Evidence Code Abbreviations:
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools





