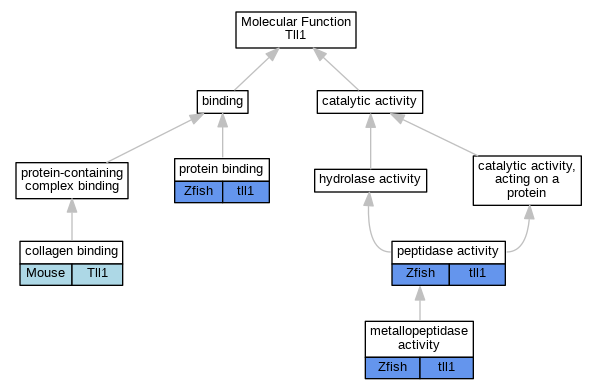
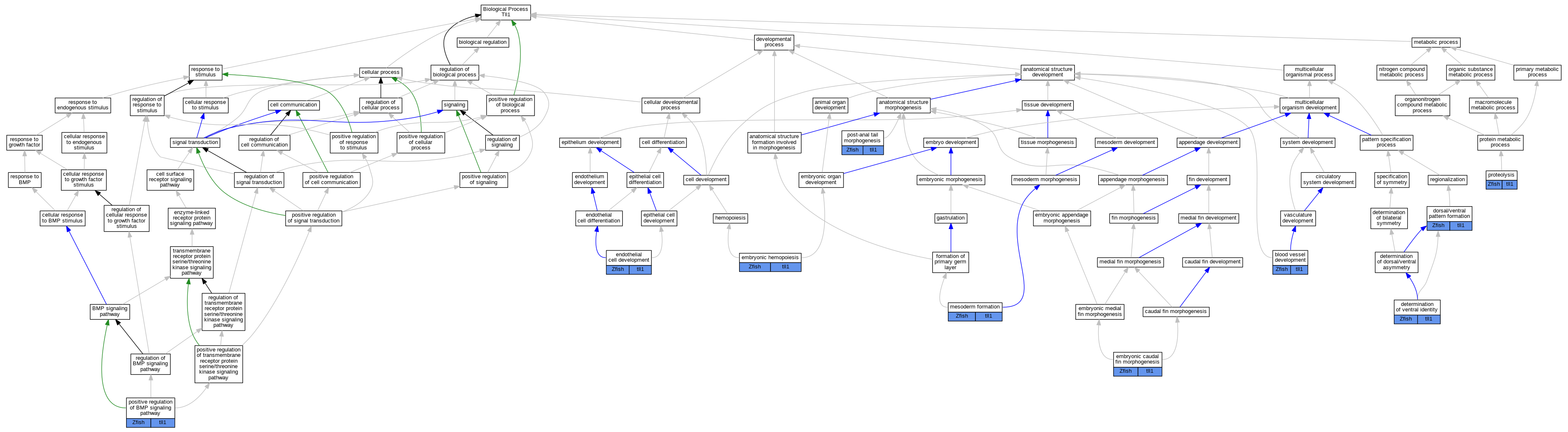
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005518 | collagen binding | Tll1 | IDA | | Mouse | PMID:12393877 |
| Molecular Function | GO:0008237 | metallopeptidase activity | tll1 | IDA | | Zfish | PMID:9395394 |
| Molecular Function | GO:0008233 | peptidase activity | tll1 | IDA | | Zfish | PMID:16518392 |
| Molecular Function | GO:0005515 | protein binding | tll1 | IPI | ZFIN:ZDB-GENE-030530-1 | Zfish | PMID:16518392 |
| Biological Process | GO:0001568 | blood vessel development | tll1 | IMP | ZFIN:ZDB-GENO-050209-23 | Zfish | PMID:10375503 |
| Biological Process | GO:0048264 | determination of ventral identity | tll1 | IMP | | Zfish | PMID:9007231 |
| Biological Process | GO:0048264 | determination of ventral identity | tll1 | IMP | ZFIN:ZDB-GENO-050209-23 | Zfish | PMID:10375503 |
| Biological Process | GO:0048264 | determination of ventral identity | tll1 | IMP | | Zfish | PMID:9395394 |
| Biological Process | GO:0048264 | determination of ventral identity | tll1 | IGI | ZFIN:ZDB-GENE-020812-3 | Zfish | PMID:15525664 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | tll1 | IMP | ZFIN:ZDB-GENO-050209-23 | Zfish | PMID:11969259 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | tll1 | IGI | ZFIN:ZDB-MRPHLNO-041217-9,ZFIN:ZDB-MRPHLNO-060818-1 | Zfish | PMID:16518392 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | tll1 | IGI | ZFIN:ZDB-MRPHLNO-041217-9,ZFIN:ZDB-MRPHLNO-050221-6,ZFIN:ZDB-MRPHLNO-060818-1 | Zfish | PMID:20886103 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | tll1 | IGI | ZFIN:ZDB-MRPHLNO-060818-4,ZFIN:ZDB-MRPHLNO-060818-7 | Zfish | PMID:16824737 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | tll1 | IMP | ZFIN:ZDB-GENO-001220-1 | Zfish | PMID:16530746 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | tll1 | IGI | ZFIN:ZDB-MRPHLNO-041217-9,ZFIN:ZDB-MRPHLNO-060818-1 | Zfish | PMID:20886103 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | tll1 | IGI | ZFIN:ZDB-MRPHLNO-041217-9,ZFIN:ZDB-MRPHLNO-060818-1,ZFIN:ZDB-MRPHLNO-060818-2 | Zfish | PMID:16518392 |
| Biological Process | GO:0035124 | embryonic caudal fin morphogenesis | tll1 | IMP | ZFIN:ZDB-GENO-050209-23 | Zfish | PMID:10375503 |
| Biological Process | GO:0035124 | embryonic caudal fin morphogenesis | tll1 | IMP | | Zfish | PMID:9395394 |
| Biological Process | GO:0035162 | embryonic hemopoiesis | tll1 | IMP | | Zfish | PMID:9395394 |
| Biological Process | GO:0001885 | endothelial cell development | tll1 | IMP | ZFIN:ZDB-GENO-050209-23 | Zfish | PMID:10375503 |
| Biological Process | GO:0001707 | mesoderm formation | tll1 | IMP | ZFIN:ZDB-GENO-050209-23 | Zfish | PMID:10375503 |
| Biological Process | GO:0030513 | positive regulation of BMP signaling pathway | tll1 | IMP | ZFIN:ZDB-GENO-050209-23 | Zfish | PMID:10375503 |
| Biological Process | GO:0030513 | positive regulation of BMP signaling pathway | tll1 | IMP | | Zfish | PMID:9395394 |
| Biological Process | GO:0036342 | post-anal tail morphogenesis | tll1 | IMP | ZFIN:ZDB-GENO-100601-3 | Zfish | PMID:21610036 |
| Biological Process | GO:0036342 | post-anal tail morphogenesis | tll1 | IGI | ZFIN:ZDB-MRPHLNO-060724-1,ZFIN:ZDB-MRPHLNO-060724-2,ZFIN:ZDB-MRPHLNO-060818-7 | Zfish | PMID:21610036 |
| Biological Process | GO:0036342 | post-anal tail morphogenesis | tll1 | IGI | ZFIN:ZDB-GENO-100601-3,ZFIN:ZDB-MRPHLNO-060724-1,ZFIN:ZDB-MRPHLNO-060724-2 | Zfish | PMID:21610036 |
| Biological Process | GO:0036342 | post-anal tail morphogenesis | tll1 | IGI | ZFIN:ZDB-MRPHLNO-060815-1,ZFIN:ZDB-MRPHLNO-060818-7 | Zfish | PMID:21610036 |
| Biological Process | GO:0036342 | post-anal tail morphogenesis | tll1 | IGI | ZFIN:ZDB-GENO-100601-3,ZFIN:ZDB-GENO-120430-1 | Zfish | PMID:21610036 |
| Biological Process | GO:0006508 | proteolysis | tll1 | IDA | | Zfish | PMID:16518392 |
| Biological Process | GO:0006508 | proteolysis | tll1 | IDA | | Zfish | PMID:9395394 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools