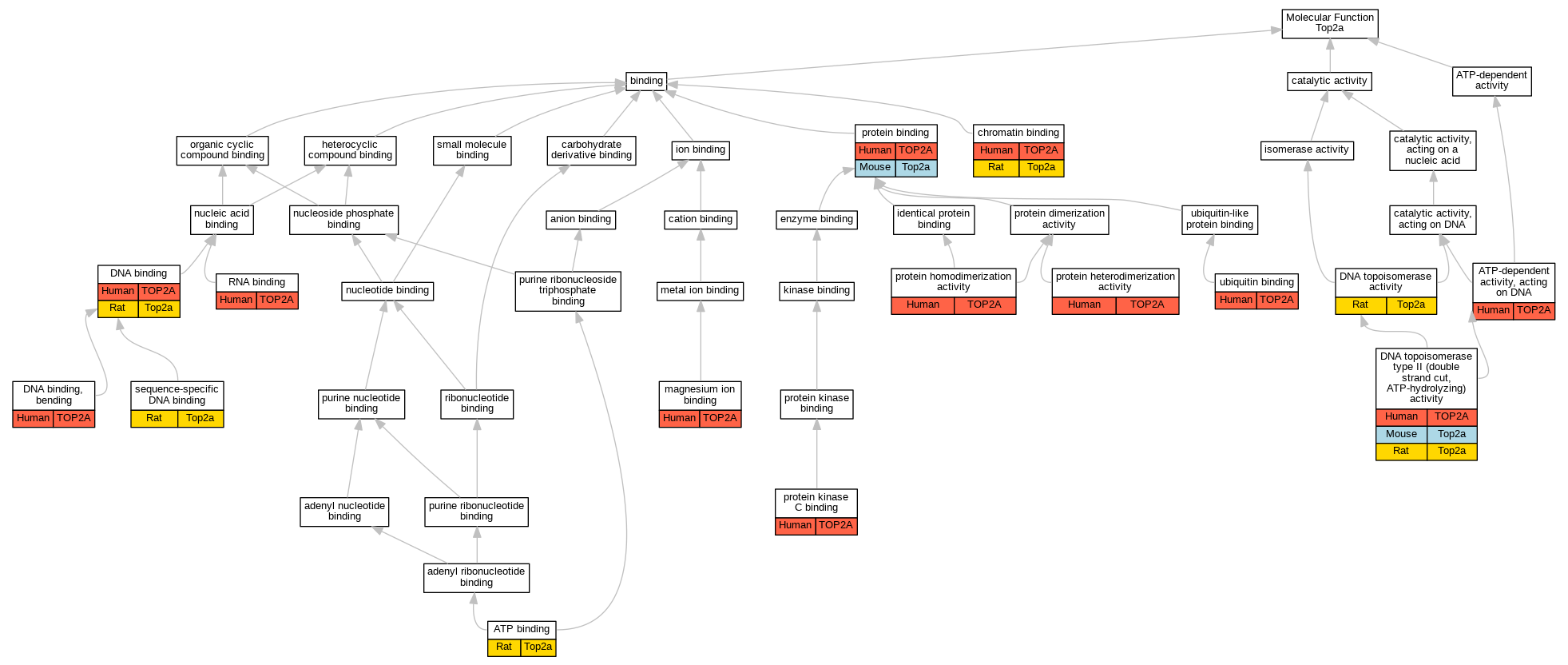
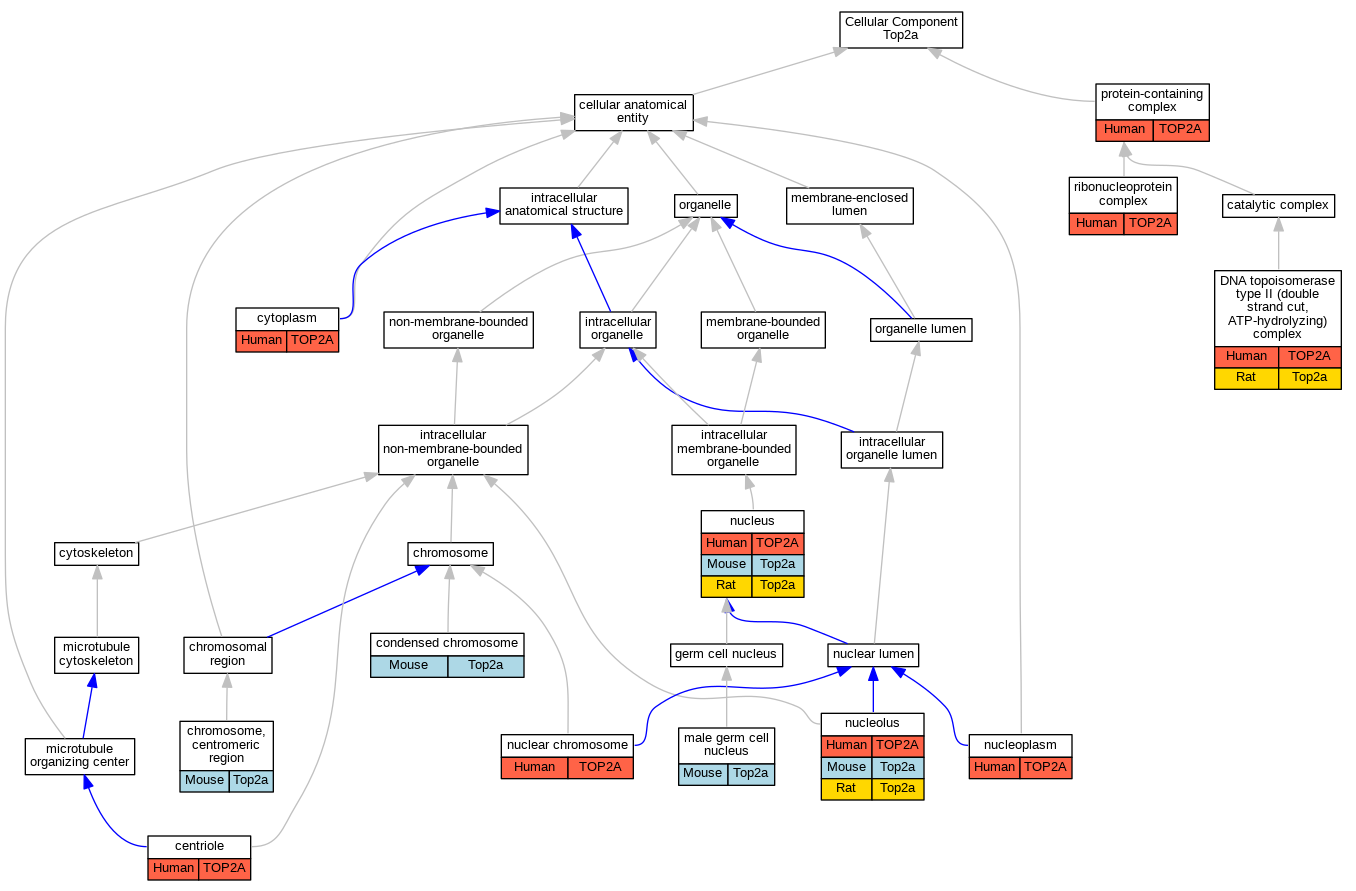
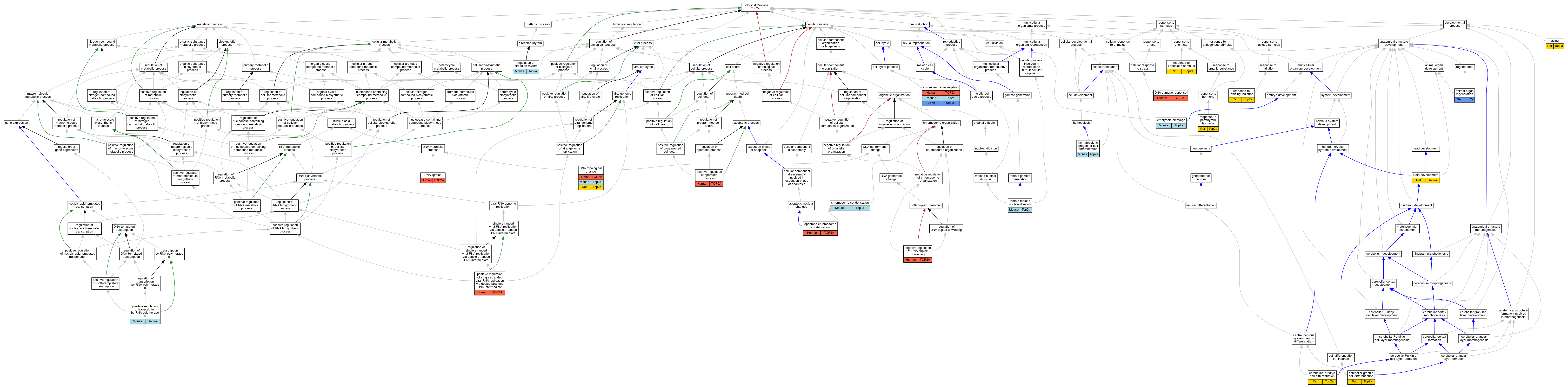
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005524 | ATP binding | Top2a | IDA | | Rat | PMID:11334111|RGD:1580614 |
| Molecular Function | GO:0008094 | ATP-dependent activity, acting on DNA | TOP2A | IDA | | Human | PMID:12079377 |
| Molecular Function | GO:0003682 | chromatin binding | TOP2A | IDA | | Human | PMID:9049244 |
| Molecular Function | GO:0003682 | chromatin binding | Top2a | IDA | | Rat | PMID:11334111|RGD:1580614 |
| Molecular Function | GO:0003677 | DNA binding | TOP2A | IDA | | Human | PMID:10788521 |
| Molecular Function | GO:0003677 | DNA binding | TOP2A | IDA | | Human | PMID:12079377 |
| Molecular Function | GO:0003677 | DNA binding | TOP2A | IDA | | Human | PMID:9049244 |
| Molecular Function | GO:0003677 | DNA binding | Top2a | IDA | | Rat | PMID:11334111|RGD:1580614 |
| Molecular Function | GO:0008301 | DNA binding, bending | TOP2A | IDA | | Human | PMID:22323612 |
| Molecular Function | GO:0003916 | DNA topoisomerase activity | Top2a | IDA | | Rat | PMID:11334111|RGD:1580614 |
| Molecular Function | GO:0003918 | DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity | TOP2A | IMP | | Human | PMID:12711669 |
| Molecular Function | GO:0003918 | DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity | TOP2A | IDA | | Human | PMID:16611985 |
| Molecular Function | GO:0003918 | DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity | Top2a | IDA | | Mouse | MGI:MGI:51613|PMID:1331984 |
| Molecular Function | GO:0003918 | DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity | Top2a | IDA | | Mouse | PMID:7842491 |
| Molecular Function | GO:0003918 | DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity | Top2a | IDA | | Mouse | PMID:9094096 |
| Molecular Function | GO:0003918 | DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity | Top2a | IDA | | Mouse | PMID:1331984 |
| Molecular Function | GO:0003918 | DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity | Top2a | IGI | SGD:S000005032 | Mouse | PMID:1331984 |
| Molecular Function | GO:0003918 | DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity | Top2a | IGI | SGD:S000005032 | Mouse | PMID:7937150 |
| Molecular Function | GO:0003918 | DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity | Top2a | IDA | | Rat | PMID:11334111|RGD:1580614 |
| Molecular Function | GO:0000287 | magnesium ion binding | TOP2A | IDA | | Human | PMID:22323612 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:P04637 | Human | PMID:10666337 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:P62258 | Human | PMID:10788521 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:O76075 | Human | PMID:10959840 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:Q13547 | Human | PMID:11062478 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:Q92769 | Human | PMID:11062478 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:Q13547 | Human | PMID:11136718 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:Q08211 | Human | PMID:12711669 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:P38398 | Human | PMID:15965487 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:Q05655 | Human | PMID:16611985 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:O15162 | Human | PMID:17567603 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:P35222 | Human | PMID:17983804 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:Q53H47 | Human | PMID:18790802 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:Q53H47 | Human | PMID:19390626 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:Q53H47 | Human | PMID:20457750 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:O60318 | Human | PMID:23652018 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:O14497-1 | Human | PMID:23698369 |
| Molecular Function | GO:0005515 | protein binding | TOP2A | IPI | UniProtKB:Q03468 | Human | PMID:26030138 |
| Molecular Function | GO:0005515 | protein binding | Top2a | IPI | UniProtKB:Q3TKT4 | Mouse | MGI:MGI:5499063|PMID:23698369 |
| Molecular Function | GO:0005515 | protein binding | Top2a | IPI | PR:A0A1D9BZF0 | Mouse | PMID:31839538 |
| Molecular Function | GO:0046982 | protein heterodimerization activity | TOP2A | IPI | UniProtKB:Q02880 | Human | PMID:10473615 |
| Molecular Function | GO:0042803 | protein homodimerization activity | TOP2A | IPI | UniProtKB:P11388 | Human | PMID:10473615 |
| Molecular Function | GO:0005080 | protein kinase C binding | TOP2A | IPI | UniProtKB:Q05655 | Human | PMID:16611985 |
| Molecular Function | GO:0003723 | RNA binding | TOP2A | HDA | | Human | PMID:22681889 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | Top2a | IDA | | Rat | PMID:11334111|RGD:1580614 |
| Molecular Function | GO:0043130 | ubiquitin binding | TOP2A | IMP | | Human | PMID:15965487 |
| Cellular Component | GO:0005814 | centriole | TOP2A | IDA | | Human | PMID:9049244 |
| Cellular Component | GO:0000775 | chromosome, centromeric region | Top2a | IDA | | Mouse | PMID:26919979 |
| Cellular Component | GO:0000793 | condensed chromosome | Top2a | IMP | | Mouse | PMID:21795393 |
| Cellular Component | GO:0000793 | condensed chromosome | Top2a | IDA | | Mouse | PMID:11835579 |
| Cellular Component | GO:0005737 | cytoplasm | TOP2A | IDA | | Human | PMID:9155056 |
| Cellular Component | GO:0009330 | DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) complex | TOP2A | IDA | | Human | PMID:10473615 |
| Cellular Component | GO:0009330 | DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) complex | Top2a | IDA | | Rat | PMID:11334111|RGD:1580614 |
| Cellular Component | GO:0001673 | male germ cell nucleus | Top2a | IDA | | Mouse | PMID:9094096 |
| Cellular Component | GO:0000228 | nuclear chromosome | TOP2A | IDA | | Human | PMID:9049244 |
| Cellular Component | GO:0005730 | nucleolus | TOP2A | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005730 | nucleolus | TOP2A | IDA | | Human | PMID:17567603 |
| Cellular Component | GO:0005730 | nucleolus | TOP2A | IDA | | Human | PMID:8299728 |
| Cellular Component | GO:0005730 | nucleolus | TOP2A | IDA | | Human | PMID:9155056 |
| Cellular Component | GO:0005730 | nucleolus | Top2a | IDA | | Mouse | PMID:11835579 |
| Cellular Component | GO:0005730 | nucleolus | Top2a | IDA | | Rat | PMID:11170002|RGD:8693630 |
| Cellular Component | GO:0005654 | nucleoplasm | TOP2A | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005654 | nucleoplasm | TOP2A | IDA | | Human | PMID:8299728 |
| Cellular Component | GO:0005654 | nucleoplasm | TOP2A | IDA | | Human | PMID:9049244 |
| Cellular Component | GO:0005634 | nucleus | TOP2A | IDA | | Human | PMID:10788521 |
| Cellular Component | GO:0005634 | nucleus | TOP2A | IDA | | Human | PMID:10959840 |
| Cellular Component | GO:0005634 | nucleus | TOP2A | IDA | | Human | PMID:16611985 |
| Cellular Component | GO:0005634 | nucleus | TOP2A | IDA | | Human | PMID:9155056 |
| Cellular Component | GO:0005634 | nucleus | Top2a | IDA | | Mouse | PMID:25961503 |
| Cellular Component | GO:0005634 | nucleus | Top2a | IDA | | Mouse | PMID:9224616 |
| Cellular Component | GO:0005634 | nucleus | Top2a | IDA | | Rat | PMID:11334111|RGD:1580614 |
| Cellular Component | GO:0005634 | nucleus | Top2a | IDA | | Rat | PMID:8390253|RGD:61772 |
| Cellular Component | GO:0032991 | protein-containing complex | TOP2A | IDA | | Human | PMID:16611985 |
| Cellular Component | GO:1990904 | ribonucleoprotein complex | TOP2A | IDA | | Human | PMID:12711669 |
| Biological Process | GO:0007568 | aging | Top2a | IEP | | Rat | PMID:19204916|RGD:2315123 |
| Biological Process | GO:0031100 | animal organ regeneration | top2a | IMP | ZFIN:ZDB-GENO-090727-2 | Zfish | PMID:19380487 |
| Biological Process | GO:0031100 | animal organ regeneration | top2a | IMP | ZFIN:ZDB-GENO-090727-1 | Zfish | PMID:19380487 |
| Biological Process | GO:0030263 | apoptotic chromosome condensation | TOP2A | IDA | | Human | PMID:10959840 |
| Biological Process | GO:0007420 | brain development | Top2a | IEP | | Rat | PMID:8008235|RGD:8693613 |
| Biological Process | GO:0021707 | cerebellar granule cell differentiation | Top2a | IEP | | Rat | PMID:11170002|RGD:8693630 |
| Biological Process | GO:0021702 | cerebellar Purkinje cell differentiation | Top2a | IEP | | Rat | PMID:11170002|RGD:8693630 |
| Biological Process | GO:0030261 | chromosome condensation | Top2a | IMP | | Mouse | PMID:11835580 |
| Biological Process | GO:0007059 | chromosome segregation | TOP2A | IMP | | Human | PMID:11136718 |
| Biological Process | GO:0007059 | chromosome segregation | TOP2A | IMP | | Human | PMID:15456904 |
| Biological Process | GO:0007059 | chromosome segregation | TOP2A | IMP | | Human | PMID:15965487 |
| Biological Process | GO:0007059 | chromosome segregation | Top2a | IMP | | Mouse | PMID:11835580 |
| Biological Process | GO:0007059 | chromosome segregation | Top2a | IDA | | Mouse | PMID:7842491 |
| Biological Process | GO:0007059 | chromosome segregation | top2a | IMP | ZFIN:ZDB-GENO-090714-1 | Zfish | PMID:19380487 |
| Biological Process | GO:0007059 | chromosome segregation | top2a | IMP | ZFIN:ZDB-MRPHLNO-090713-1 | Zfish | PMID:19380487 |
| Biological Process | GO:0006974 | DNA damage response | TOP2A | IDA | | Human | PMID:16611985 |
| Biological Process | GO:0006266 | DNA ligation | TOP2A | IDA | | Human | PMID:15491148 |
| Biological Process | GO:0006265 | DNA topological change | TOP2A | IMP | | Human | PMID:12711669 |
| Biological Process | GO:0006265 | DNA topological change | TOP2A | IDA | | Human | PMID:22323612 |
| Biological Process | GO:0006265 | DNA topological change | Top2a | IDA | | Mouse | PMID:9224616 |
| Biological Process | GO:0006265 | DNA topological change | Top2a | IDA | | Mouse | PMID:1331984 |
| Biological Process | GO:0006265 | DNA topological change | Top2a | IDA | | Rat | PMID:11334111|RGD:1580614 |
| Biological Process | GO:0040016 | embryonic cleavage | Top2a | IMP | | Mouse | PMID:11835580 |
| Biological Process | GO:0007143 | female meiotic nuclear division | Top2a | IMP | | Mouse | PMID:25961503 |
| Biological Process | GO:0002244 | hematopoietic progenitor cell differentiation | Top2a | IMP | | Mouse | PMID:24029230 |
| Biological Process | GO:1905463 | negative regulation of DNA duplex unwinding | TOP2A | IMP | | Human | PMID:12711669 |
| Biological Process | GO:0043065 | positive regulation of apoptotic process | TOP2A | IDA | | Human | PMID:16611985 |
| Biological Process | GO:0045870 | positive regulation of single stranded viral RNA replication via double stranded DNA intermediate | TOP2A | IMP | | Human | PMID:16712776 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Top2a | IMP | | Mouse | PMID:1683482 |
| Biological Process | GO:0042752 | regulation of circadian rhythm | Top2a | IMP | | Mouse | MGI:MGI:5613450|PMID:24321095 |
| Biological Process | GO:0010212 | response to ionizing radiation | Top2a | IEP | | Rat | PMID:22965249|RGD:13432139 |
| Biological Process | GO:0071107 | response to parathyroid hormone | Top2a | IEP | | Rat | PMID:10709994|RGD:2315136 |
| Biological Process | GO:0009410 | response to xenobiotic stimulus | Top2a | IEP | | Rat | PMID:14766721|RGD:2315135 |
| Biological Process | GO:0009410 | response to xenobiotic stimulus | Top2a | IEP | | Rat | PMID:15033592|RGD:2315134 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools