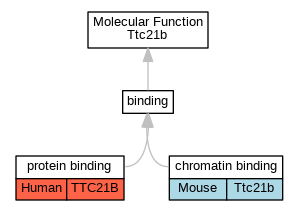
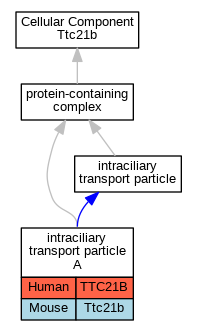
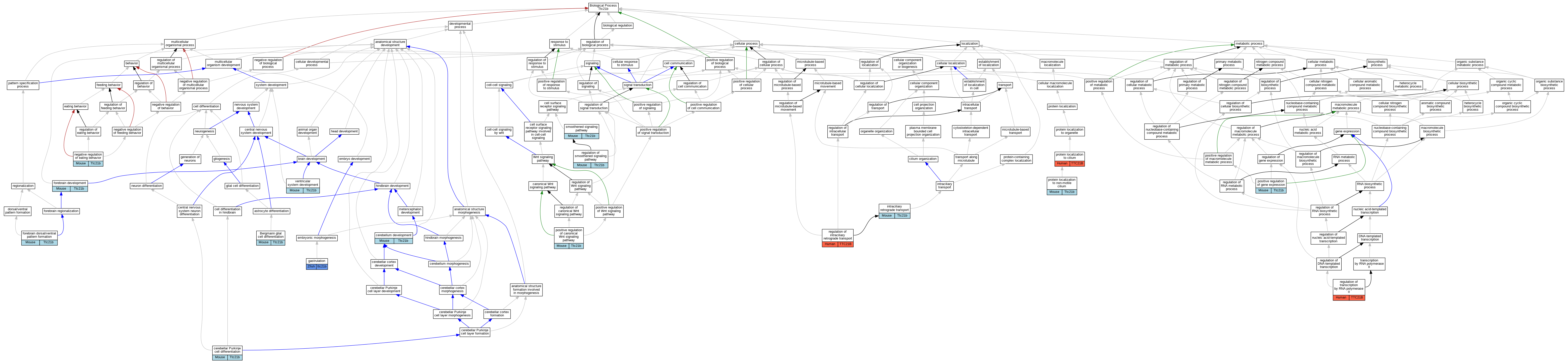
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0003682 | chromatin binding | Ttc21b | IDA | | Mouse | PMID:22079090 |
| Molecular Function | GO:0005515 | protein binding | TTC21B | IPI | UniProtKB:Q9HBG6-3 | Human | PMID:29220510 |
| Cellular Component | GO:0030991 | intraciliary transport particle A | TTC21B | IPI | | Human | PMID:27173435 |
| Cellular Component | GO:0030991 | intraciliary transport particle A | TTC21B | IDA | | Human | PMID:20889716 |
| Cellular Component | GO:0030991 | intraciliary transport particle A | TTC21B | IDA | | Human | PMID:27932497 |
| Cellular Component | GO:0030991 | intraciliary transport particle A | Ttc21b | IDA | | Mouse | PMID:20889716 |
| Biological Process | GO:0060020 | Bergmann glial cell differentiation | Ttc21b | IMP | MGI:MGI:2152544,MGI:MGI:2179048,MGI:MGI:5587034 | Mouse | PMID:29615573 |
| Biological Process | GO:0021702 | cerebellar Purkinje cell differentiation | Ttc21b | IMP | MGI:MGI:2152544,MGI:MGI:2179048,MGI:MGI:5587034 | Mouse | PMID:29615573 |
| Biological Process | GO:0021702 | cerebellar Purkinje cell differentiation | Ttc21b | IMP | MGI:MGI:2137515,MGI:MGI:2152544,MGI:MGI:5587034 | Mouse | PMID:29615573 |
| Biological Process | GO:0021549 | cerebellum development | Ttc21b | IMP | MGI:MGI:2152544,MGI:MGI:2179048,MGI:MGI:5587034 | Mouse | PMID:29615573 |
| Biological Process | GO:0030900 | forebrain development | Ttc21b | IMP | MGI:MGI:2686837,MGI:MGI:5587034 | Mouse | PMID:28291836 |
| Biological Process | GO:0021798 | forebrain dorsal/ventral pattern formation | Ttc21b | IMP | MGI:MGI:2152544 | Mouse | PMID:19732765 |
| Biological Process | GO:0007369 | gastrulation | ttc21b | IMP | ZFIN:ZDB-MRPHLNO-141007-2 | Zfish | PMID:21258341 |
| Biological Process | GO:0007369 | gastrulation | ttc21b | IMP | ZFIN:ZDB-MRPHLNO-141007-1 | Zfish | PMID:21258341 |
| Biological Process | GO:0035721 | intraciliary retrograde transport | Ttc21b | IMP | | Mouse | PMID:18327258 |
| Biological Process | GO:0035721 | intraciliary retrograde transport | Ttc21b | IMP | MGI:MGI:2152544,MGI:MGI:2669525,MGI:MGI:5587034 | Mouse | PMID:27482817 |
| Biological Process | GO:0035721 | intraciliary retrograde transport | Ttc21b | IMP | MGI:MGI:2152544 | Mouse | PMID:19732765 |
| Biological Process | GO:1903999 | negative regulation of eating behavior | Ttc21b | IMP | MGI:MGI:2152544,MGI:MGI:2669525,MGI:MGI:5587034 | Mouse | PMID:27482817 |
| Biological Process | GO:0090263 | positive regulation of canonical Wnt signaling pathway | Ttc21b | IMP | MGI:MGI:2152544,MGI:MGI:2669525,MGI:MGI:5587034 | Mouse | PMID:27482817 |
| Biological Process | GO:0010628 | positive regulation of gene expression | Ttc21b | IMP | MGI:MGI:2152544,MGI:MGI:2669525,MGI:MGI:5587034 | Mouse | PMID:27482817 |
| Biological Process | GO:0061512 | protein localization to cilium | TTC21B | IMP | | Human | PMID:20889716 |
| Biological Process | GO:0097499 | protein localization to non-motile cilium | Ttc21b | IMP | MGI:MGI:2152544,MGI:MGI:2669525,MGI:MGI:5587034 | Mouse | PMID:27482817 |
| Biological Process | GO:1905799 | regulation of intraciliary retrograde transport | TTC21B | IMP | | Human | PMID:27932497 |
| Biological Process | GO:0008589 | regulation of smoothened signaling pathway | Ttc21b | IMP | MGI:MGI:2152544 | Mouse | PMID:19732765 |
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | TTC21B | IMP | | Human | PMID:22302990 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ttc21b | IMP | MGI:MGI:2152544,MGI:MGI:2179048,MGI:MGI:5587034 | Mouse | PMID:29615573 |
| Biological Process | GO:0007224 | smoothened signaling pathway | Ttc21b | IMP | MGI:MGI:2152544 | Mouse | PMID:18327258 |
| Biological Process | GO:0021591 | ventricular system development | Ttc21b | IMP | MGI:MGI:2152544 | Mouse | PMID:19732765 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools