|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
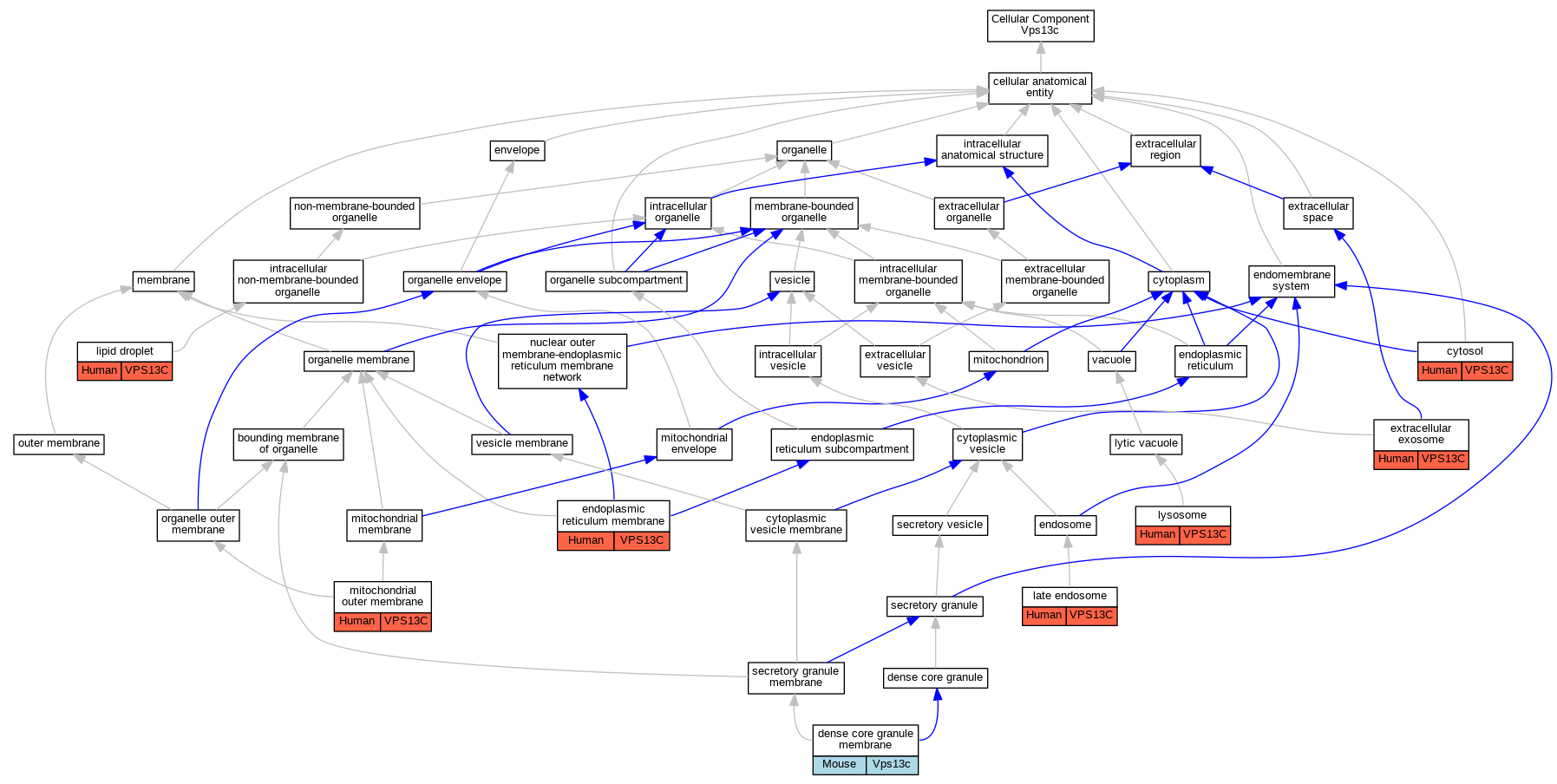
| Cellular Component | GO:0005829 | cytosol | VPS13C | IDA | Human | PMID:26942284 | |
| Cellular Component | GO:0032127 | dense core granule membrane | Vps13c | IDA | Mouse | PMID:27329800 | |
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | VPS13C | IDA | Human | PMID:30093493 | |
| Cellular Component | GO:0070062 | extracellular exosome | VPS13C | HDA | Human | PMID:19056867 | |
| Cellular Component | GO:0005770 | late endosome | VPS13C | IDA | Human | PMID:30093493 | |
| Cellular Component | GO:0005811 | lipid droplet | VPS13C | IDA | Human | PMID:30093493 | |
| Cellular Component | GO:0005764 | lysosome | VPS13C | IDA | Human | PMID:30093493 | |
| Cellular Component | GO:0005741 | mitochondrial outer membrane | VPS13C | IDA | Human | PMID:26942284 | |
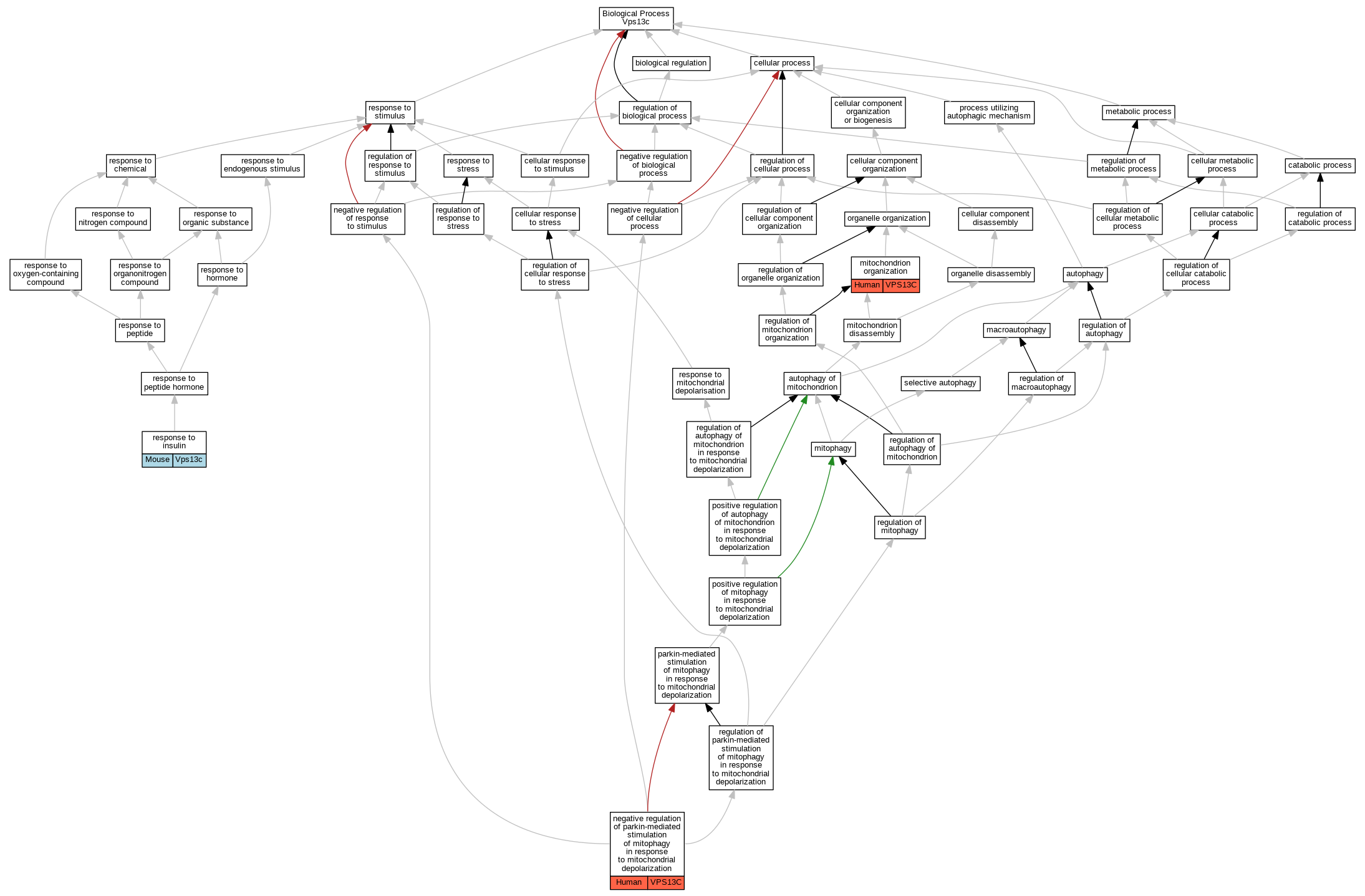
| Biological Process | GO:0007005 | mitochondrion organization | VPS13C | IMP | Human | PMID:26942284 | |
| Biological Process | GO:1905090 | negative regulation of parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | VPS13C | IMP | Human | PMID:26942284 | |
| Biological Process | GO:0032868 | response to insulin | Vps13c | IMP | MGI:MGI:4410453,MGI:MGI:5907128 | Mouse | PMID:27329800 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 07/02/2024 MGI 6.13 |

|
|
|
||