|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
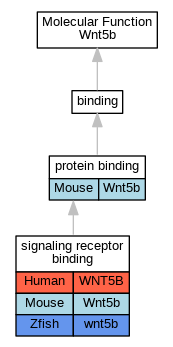
| Molecular Function | GO:0005515 | protein binding | Wnt5b | IPI | UniProtKB:Q9CWT0 | Mouse | PMID:10866835 |
| Molecular Function | GO:0005102 | signaling receptor binding | WNT5B | IPI | UniProtKB:Q01974 | Human | PMID:19486338 |
| Molecular Function | GO:0005102 | signaling receptor binding | Wnt5b | IPI | UniProtKB:Q61091 | Mouse | MGI:MGI:5706913|PMID:20093360 |
| Molecular Function | GO:0005102 | signaling receptor binding | wnt5b | IPI | ZFIN:ZDB-GENE-070209-277 | Zfish | PMID:20660632 |
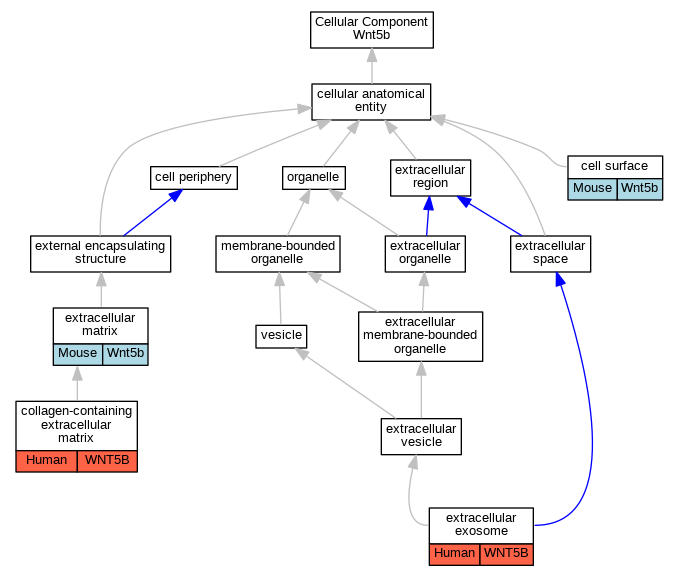
| Cellular Component | GO:0009986 | cell surface | Wnt5b | IDA | Mouse | MGI:MGI:66576|PMID:8167409 | |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | WNT5B | HDA | Human | PMID:28327460 | |
| Cellular Component | GO:0070062 | extracellular exosome | WNT5B | HDA | Human | PMID:19056867 | |
| Cellular Component | GO:0031012 | extracellular matrix | Wnt5b | IDA | Mouse | PMID:8167409 | |
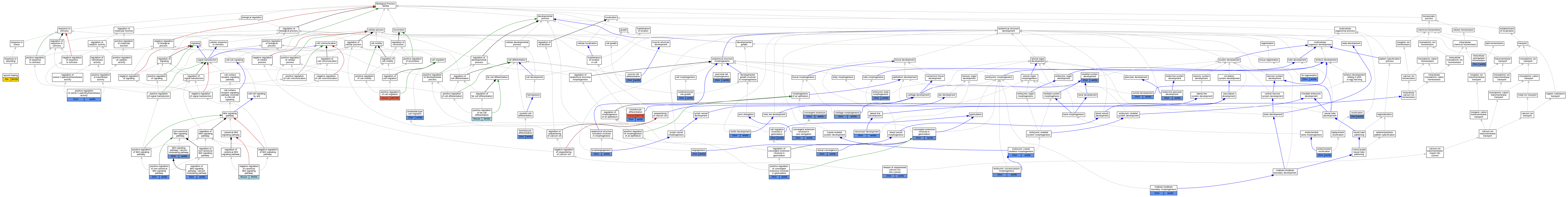
| Biological Process | GO:0001667 | ameboidal-type cell migration | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-051207-1 | Zfish | PMID:16246260 |
| Biological Process | GO:0001525 | angiogenesis | wnt5b | IMP | ZFIN:ZDB-GENO-090915-2 | Zfish | PMID:18798004 |
| Biological Process | GO:0001525 | angiogenesis | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-090915-1 | Zfish | PMID:18798004 |
| Biological Process | GO:0060536 | cartilage morphogenesis | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1367 | Zfish | PMID:26459057 |
| Biological Process | GO:0060536 | cartilage morphogenesis | wnt5b | IMP | ZFIN:ZDB-GENO-180116-3 | Zfish | PMID:27908786 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | wnt5b | IGI | ZFIN:ZDB-MRPHLNO-041217-10,ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:17032765 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:16019189 |
| Biological Process | GO:0002062 | chondrocyte differentiation | WNT5B | IEP | Human | PMID:15135146 | |
| Biological Process | GO:0002062 | chondrocyte differentiation | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1366 | Zfish | PMID:9007254 |
| Biological Process | GO:0002062 | chondrocyte differentiation | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1002 | Zfish | PMID:9007254 |
| Biological Process | GO:0002062 | chondrocyte differentiation | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1370 | Zfish | PMID:9007254 |
| Biological Process | GO:0002062 | chondrocyte differentiation | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1367 | Zfish | PMID:9007254 |
| Biological Process | GO:0060026 | convergent extension | wnt5b | IGI | ZFIN:ZDB-GENO-080222-3,ZFIN:ZDB-MRPHLNO-060126-1 | Zfish | PMID:17906624 |
| Biological Process | GO:0060026 | convergent extension | wnt5b | IGI | ZFIN:ZDB-GENO-080222-3,ZFIN:ZDB-MRPHLNO-060208-1 | Zfish | PMID:17906624 |
| Biological Process | GO:0060026 | convergent extension | wnt5b | IGI | ZFIN:ZDB-GENO-080222-3,ZFIN:ZDB-MRPHLNO-060208-2 | Zfish | PMID:17906624 |
| Biological Process | GO:0060028 | convergent extension involved in axis elongation | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-090915-1 | Zfish | PMID:20660632 |
| Biological Process | GO:0060028 | convergent extension involved in axis elongation | wnt5b | IGI | ZFIN:ZDB-MRPHLNO-090915-1,ZFIN:ZDB-MRPHLNO-100805-1 | Zfish | PMID:20660632 |
| Biological Process | GO:0060028 | convergent extension involved in axis elongation | wnt5b | IGI | ZFIN:ZDB-GENE-030827-4 | Zfish | PMID:22100263 |
| Biological Process | GO:0060028 | convergent extension involved in axis elongation | wnt5b | IGI | ZFIN:ZDB-GENO-080512-4 | Zfish | PMID:14660439 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | wnt5b | IGI | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:25183869 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | wnt5b | IGI | ZFIN:ZDB-MRPHLNO-041217-11,ZFIN:ZDB-MRPHLNO-120614-3 | Zfish | PMID:22357957 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | wnt5b | IGI | ZFIN:ZDB-MRPHLNO-041217-11,ZFIN:ZDB-MRPHLNO-080801-1 | Zfish | PMID:18159945 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:18159945 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:12676324 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | wnt5b | IGI | ZFIN:ZDB-MRPHLNO-041217-11,ZFIN:ZDB-MRPHLNO-050128-3,ZFIN:ZDB-MRPHLNO-070304-3 | Zfish | PMID:15815683 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:15815683 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | wnt5b | IMP | ZFIN:ZDB-GENO-100614-2 | Zfish | PMID:12676324 |
| Biological Process | GO:0060030 | dorsal convergence | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-090915-1,ZFIN:ZDB-MRPHLNO-100805-1 | Zfish | PMID:20660632 |
| Biological Process | GO:0010172 | embryonic body morphogenesis | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1367 | Zfish | PMID:9598355 |
| Biological Process | GO:0010172 | embryonic body morphogenesis | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1371 | Zfish | PMID:9598355 |
| Biological Process | GO:0048701 | embryonic cranial skeleton morphogenesis | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1367 | Zfish | PMID:9007236 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1370 | Zfish | PMID:25934698 |
| Biological Process | GO:0001958 | endochondral ossification | wnt5b | IMP | ZFIN:ZDB-GENO-180116-3 | Zfish | PMID:27908786 |
| Biological Process | GO:0031018 | endocrine pancreas development | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-051207-1 | Zfish | PMID:16246260 |
| Biological Process | GO:0031101 | fin regeneration | wnt5b | IMP | Zfish | PMID:19445916 | |
| Biological Process | GO:0031101 | fin regeneration | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1367 | Zfish | PMID:17185322 |
| Biological Process | GO:0090156 | intracellular sphingolipid homeostasis | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:32620775 |
| Biological Process | GO:0001946 | lymphangiogenesis | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:25992545 |
| Biological Process | GO:0001946 | lymphangiogenesis | wnt5b | IMP | ZFIN:ZDB-GENO-151110-11 | Zfish | PMID:25992545 |
| Biological Process | GO:0021555 | midbrain-hindbrain boundary morphogenesis | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-051207-1 | Zfish | PMID:30305282 |
| Biological Process | GO:0021555 | midbrain-hindbrain boundary morphogenesis | wnt5b | IGI | ZFIN:ZDB-GENE-020114-1 | Zfish | PMID:30305282 |
| Biological Process | GO:0042692 | muscle cell differentiation | wnt5b | IMP | Zfish | PMID:22406073|ZFIN:ZDB-PUB-120314-20 | |
| Biological Process | GO:0090090 | negative regulation of canonical Wnt signaling pathway | Wnt5b | IGI | MGI:MGI:98956 | Mouse | PMID:15796911 |
| Biological Process | GO:0048884 | neuromast development | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:22308195 |
| Biological Process | GO:0001503 | ossification | wnt5b | IMP | ZFIN:ZDB-GENO-180116-3 | Zfish | PMID:27908786 |
| Biological Process | GO:0048840 | otolith development | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:22308195 |
| Biological Process | GO:0030335 | positive regulation of cell migration | WNT5B | IDA | Human | PMID:19486338 | |
| Biological Process | GO:1904105 | positive regulation of convergent extension involved in gastrulation | wnt5b | IMP | Zfish | PMID:22406073|ZFIN:ZDB-PUB-120314-20 | |
| Biological Process | GO:0045600 | positive regulation of fat cell differentiation | Wnt5b | IGI | MGI:MGI:98956 | Mouse | PMID:15796911 |
| Biological Process | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway | wnt5b | IMP | Zfish | PMID:22406073|ZFIN:ZDB-PUB-120314-20 | |
| Biological Process | GO:1904222 | positive regulation of serine C-palmitoyltransferase activity | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:32620775 |
| Biological Process | GO:0036342 | post-anal tail morphogenesis | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1367 | Zfish | PMID:9007236 |
| Biological Process | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway | wnt5b | IGI | ZFIN:ZDB-MRPHLNO-110523-7 | Zfish | PMID:20628572 |
| Biological Process | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-090915-1 | Zfish | PMID:20628572 |
| Biological Process | GO:0051209 | release of sequestered calcium ion into cytosol | wnt5b | IDA | Zfish | PMID:20660632 | |
| Biological Process | GO:0001501 | skeletal system development | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1371 | Zfish | PMID:9598355 |
| Biological Process | GO:0001501 | skeletal system development | wnt5b | IMP | ZFIN:ZDB-GENO-980202-1367 | Zfish | PMID:9598355 |
| Biological Process | GO:0061053 | somite development | wnt5b | IGI | ZFIN:ZDB-MRPHLNO-110523-7 | Zfish | PMID:20628572 |
| Biological Process | GO:0002574 | thrombocyte differentiation | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:22308195 |
| Biological Process | GO:0009826 | unidimensional cell growth | wnt5b | IMP | ZFIN:ZDB-MRPHLNO-041217-11 | Zfish | PMID:12676324 |
| Biological Process | GO:0009826 | unidimensional cell growth | wnt5b | IMP | ZFIN:ZDB-GENO-100614-2 | Zfish | PMID:12676324 |
| Biological Process | GO:0007223 | Wnt signaling pathway, calcium modulating pathway | wnt5b | IGI | ZFIN:ZDB-MRPHLNO-051207-1,ZFIN:ZDB-MRPHLNO-051207-2,ZFIN:ZDB-MRPHLNO-051207-3 | Zfish | PMID:16246260 |
| Biological Process | GO:0007223 | Wnt signaling pathway, calcium modulating pathway | wnt5b | IDA | Zfish | PMID:20660632 | |
| Biological Process | GO:0042060 | wound healing | Wnt5b | IEP | Rat | PMID:17001188|RGD:2317897 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 06/12/2024 MGI 6.13 |

|
|
|
||