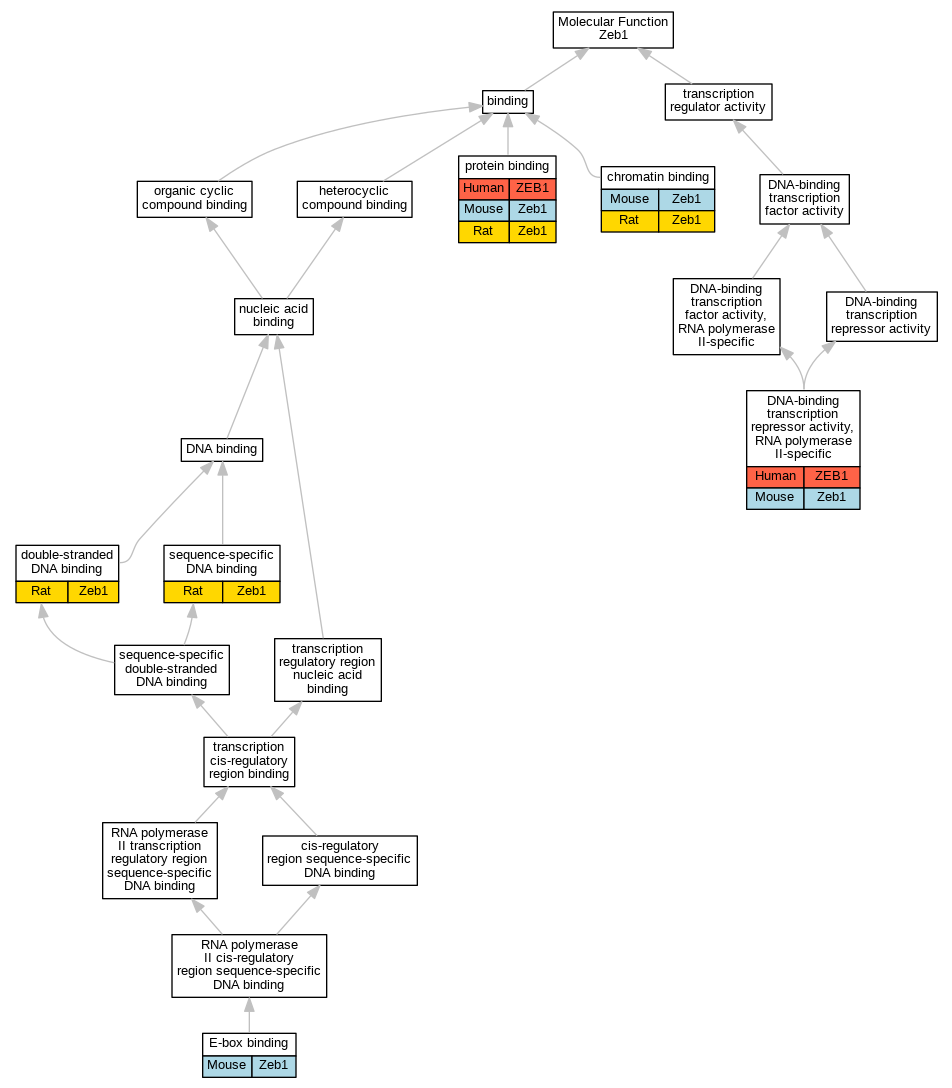
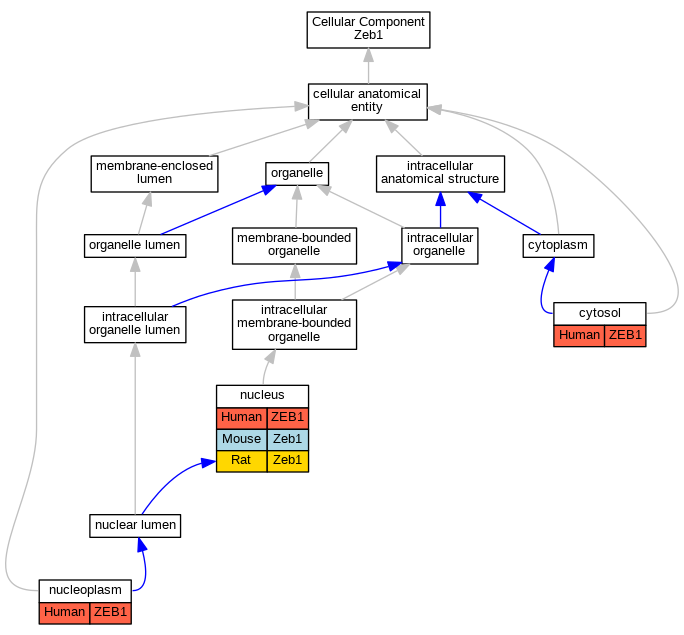
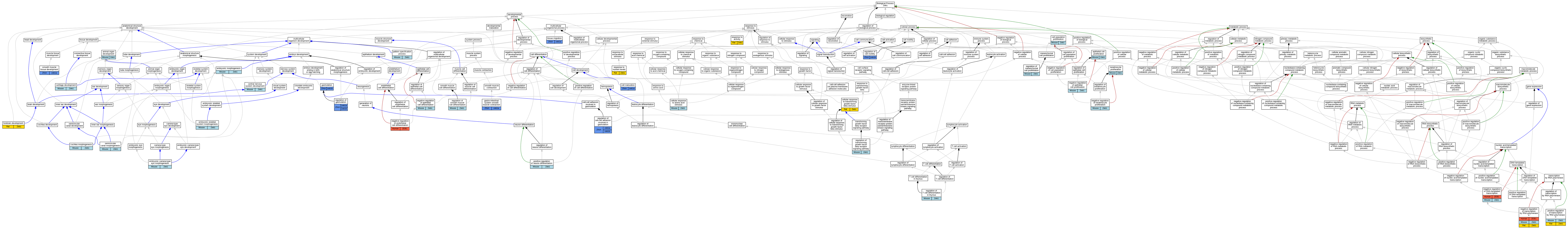
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0003682 | chromatin binding | Zeb1 | IDA | | Mouse | PMID:17392792 |
| Molecular Function | GO:0003682 | chromatin binding | Zeb1 | IDA | | Mouse | PMID:16824956 |
| Molecular Function | GO:0003682 | chromatin binding | Zeb1 | IDA | | Rat | PMID:16824956|RGD:2325877 |
| Molecular Function | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | ZEB1 | IDA | | Human | PMID:23765923 |
| Molecular Function | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | ZEB1 | IDA | | Human | PMID:23814079 |
| Molecular Function | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | Zeb1 | IDA | | Mouse | PMID:24735878 |
| Molecular Function | GO:0003690 | double-stranded DNA binding | Zeb1 | IDA | | Rat | PMID:8769566|RGD:730026 |
| Molecular Function | GO:0070888 | E-box binding | Zeb1 | IDA | | Mouse | MGI:MGI:4830798|PMID:20346398 |
| Molecular Function | GO:0005515 | protein binding | ZEB1 | IPI | UniProtKB:P51532 | Human | PMID:20418909 |
| Molecular Function | GO:0005515 | protein binding | Zeb1 | IPI | UniProtKB:O88712 | Mouse | MGI:MGI:4939879|PMID:21224849 |
| Molecular Function | GO:0005515 | protein binding | Zeb1 | IPI | PR:Q6ZQ88 | Mouse | PMID:17392792 |
| Molecular Function | GO:0005515 | protein binding | Zeb1 | IPI | PR:Q9JM73 | Mouse | PMID:16824956 |
| Molecular Function | GO:0005515 | protein binding | Zeb1 | IPI | RGD:3032 | Rat | PMID:16824956|RGD:2325877 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | Zeb1 | IDA | | Rat | PMID:15226419|RGD:2325878 |
| Cellular Component | GO:0005829 | cytosol | ZEB1 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005654 | nucleoplasm | ZEB1 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005634 | nucleus | ZEB1 | IDA | | Human | PMID:20418909 |
| Cellular Component | GO:0005634 | nucleus | ZEB1 | IDA | | Human | PMID:23814079 |
| Cellular Component | GO:0005634 | nucleus | ZEB1 | IDA | | Human | PMID:25190660 |
| Cellular Component | GO:0005634 | nucleus | Zeb1 | IDA | | Mouse | MGI:MGI:4830798|PMID:20346398 |
| Cellular Component | GO:0005634 | nucleus | Zeb1 | IDA | | Mouse | PMID:14643678 |
| Cellular Component | GO:0005634 | nucleus | Zeb1 | IDA | | Rat | PMID:15226419|RGD:2325878 |
| Biological Process | GO:0048513 | animal organ development | Zeb1 | IMP | MGI:MGI:1861230 | Mouse | PMID:11681996 |
| Biological Process | GO:0051216 | cartilage development | Zeb1 | IMP | MGI:MGI:1857799 | Mouse | PMID:9389660 |
| Biological Process | GO:0048469 | cell maturation | zeb1a | IMP | ZFIN:ZDB-MRPHLNO-170327-1 | Zfish | PMID:28049660 |
| Biological Process | GO:0008283 | cell population proliferation | Zeb1 | IMP | MGI:MGI:1857515 | Mouse | PMID:18436818 |
| Biological Process | GO:0071230 | cellular response to amino acid stimulus | Zeb1 | IDA | | Mouse | PMID:20548288 |
| Biological Process | GO:0071560 | cellular response to transforming growth factor beta stimulus | Zeb1 | IEP | | Rat | PMID:16824956|RGD:2325877 |
| Biological Process | GO:0007417 | central nervous system development | Zeb1 | IGI | MGI:MGI:1344407 | Mouse | PMID:16598713 |
| Biological Process | GO:0090103 | cochlea morphogenesis | Zeb1 | IMP | MGI:MGI:1861230 | Mouse | PMID:21980308 |
| Biological Process | GO:0048596 | embryonic camera-type eye morphogenesis | Zeb1 | IMP | MGI:MGI:1857515 | Mouse | PMID:18436818 |
| Biological Process | GO:0048598 | embryonic morphogenesis | Zeb1 | IGI | MGI:MGI:1344407 | Mouse | PMID:16598713 |
| Biological Process | GO:0048704 | embryonic skeletal system morphogenesis | Zeb1 | IMP | MGI:MGI:1857799 | Mouse | PMID:9389660 |
| Biological Process | GO:0050673 | epithelial cell proliferation | Zeb1 | IMP | MGI:MGI:1857515 | Mouse | PMID:18436818 |
| Biological Process | GO:0030900 | forebrain development | Zeb1 | IEP | | Rat | PMID:11731009|RGD:2325879 |
| Biological Process | GO:0014831 | gastro-intestinal system smooth muscle contraction | zeb1a | IMP | ZFIN:ZDB-MRPHLNO-170327-1 | Zfish | PMID:28049660 |
| Biological Process | GO:0007369 | gastrulation | zeb1b | IMP | ZFIN:ZDB-MRPHLNO-130910-1 | Zfish | PMID:20036349 |
| Biological Process | GO:0043616 | keratinocyte proliferation | Zeb1 | IMP | MGI:MGI:1857515 | Mouse | PMID:18436818 |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ZEB1 | IMP | | Human | PMID:12937339 |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ZEB1 | IDA | | Human | PMID:20418909 |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | Zeb1 | IDA | | Mouse | MGI:MGI:4830798|PMID:20346398 |
| Biological Process | GO:0045602 | negative regulation of endothelial cell differentiation | ZEB1 | IMP | | Human | PMID:23765923 |
| Biological Process | GO:0030857 | negative regulation of epithelial cell differentiation | Zeb1 | IMP | | Mouse | PMID:18192284 |
| Biological Process | GO:0050680 | negative regulation of epithelial cell proliferation | Zeb1 | IMP | MGI:MGI:1857515 | Mouse | PMID:18436818 |
| Biological Process | GO:0010839 | negative regulation of keratinocyte proliferation | Zeb1 | IMP | MGI:MGI:1857515 | Mouse | PMID:18436818 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ZEB1 | IDA | | Human | PMID:23765923 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ZEB1 | IDA | | Human | PMID:23814079 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | Zeb1 | IMP | | Mouse | PMID:17392792 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | Zeb1 | IDA | | Rat | PMID:11476947|RGD:2325880 |
| Biological Process | GO:0007389 | pattern specification process | Zeb1 | IMP | MGI:MGI:1857799 | Mouse | PMID:9389660 |
| Biological Process | GO:0045666 | positive regulation of neuron differentiation | Zeb1 | IDA | | Mouse | MGI:MGI:4830798|PMID:20346398 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Zeb1 | IDA | | Mouse | PMID:16824956 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Zeb1 | IGI | MGI:MGI:1201674 | Mouse | PMID:16824956 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Zeb1 | IGI | MGI:MGI:106658 | Mouse | PMID:16824956 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Zeb1 | IGI | MGI:MGI:894293 | Mouse | PMID:16824956 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Zeb1 | IMP | | Rat | PMID:16824956|RGD:2325877 |
| Biological Process | GO:2000145 | regulation of cell motility | zeb1b | IGI | ZFIN:ZDB-MRPHLNO-050413-1,ZFIN:ZDB-MRPHLNO-120823-1,ZFIN:ZDB-MRPHLNO-120823-2,ZFIN:ZDB-MRPHLNO-120823-3,ZFIN:ZDB-MRPHLNO-120823-4,ZFIN:ZDB-MRPHLNO-120823-5 | Zfish | PMID:22705393 |
| Biological Process | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation | zeb1b | IGI | ZFIN:ZDB-MRPHLNO-130801-1,ZFIN:ZDB-MRPHLNO-130801-2 | Zfish | PMID:23667256 |
| Biological Process | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation | zeb1a | IGI | ZFIN:ZDB-MRPHLNO-130801-1,ZFIN:ZDB-MRPHLNO-130801-2 | Zfish | PMID:23667256 |
| Biological Process | GO:0010470 | regulation of gastrulation | zeb1b | IGI | ZFIN:ZDB-MRPHLNO-130801-1,ZFIN:ZDB-MRPHLNO-130801-2 | Zfish | PMID:23667256 |
| Biological Process | GO:0010470 | regulation of gastrulation | zeb1a | IGI | ZFIN:ZDB-MRPHLNO-130801-1,ZFIN:ZDB-MRPHLNO-130801-2 | Zfish | PMID:23667256 |
| Biological Process | GO:0010464 | regulation of mesenchymal cell proliferation | Zeb1 | IMP | | Mouse | PMID:18192284 |
| Biological Process | GO:0051150 | regulation of smooth muscle cell differentiation | Zeb1 | IMP | | Mouse | PMID:16824956 |
| Biological Process | GO:0033081 | regulation of T cell differentiation in thymus | Zeb1 | IMP | MGI:MGI:1857799 | Mouse | PMID:9389660 |
| Biological Process | GO:0033081 | regulation of T cell differentiation in thymus | Zeb1 | IMP | MGI:MGI:1857515 | Mouse | PMID:9126927 |
| Biological Process | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | Zeb1 | IGI | MGI:MGI:1201674 | Mouse | PMID:16824956 |
| Biological Process | GO:0014823 | response to activity | Zeb1 | IEP | | Rat | PMID:19366873|RGD:2325875 |
| Biological Process | GO:0031667 | response to nutrient levels | Zeb1 | IEP | | Rat | PMID:19366873|RGD:2325875 |
| Biological Process | GO:0048752 | semicircular canal morphogenesis | Zeb1 | IMP | MGI:MGI:1861230 | Mouse | PMID:21980308 |
| Biological Process | GO:0048745 | smooth muscle tissue development | zeb1a | IMP | ZFIN:ZDB-MRPHLNO-170327-1 | Zfish | PMID:28049660 |
| Biological Process | GO:0090130 | tissue migration | zeb1a | IMP | ZFIN:ZDB-MRPHLNO-170327-1 | Zfish | PMID:28049660 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools