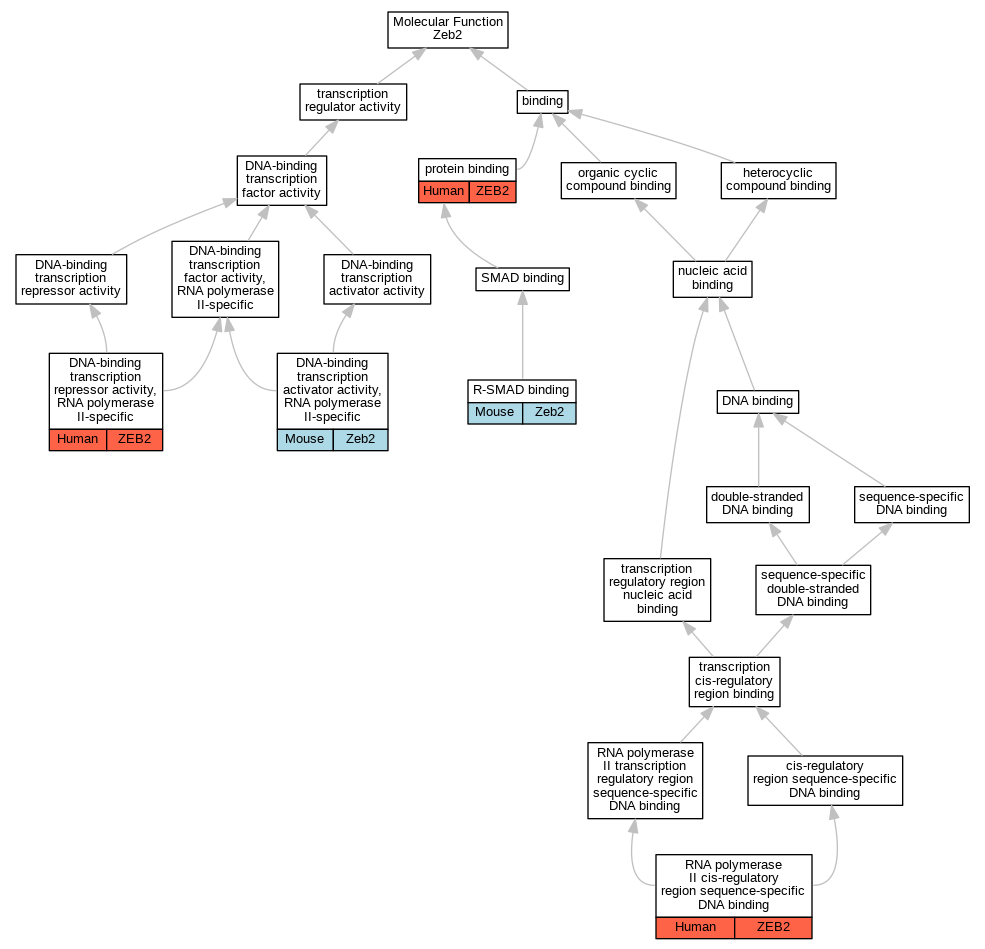
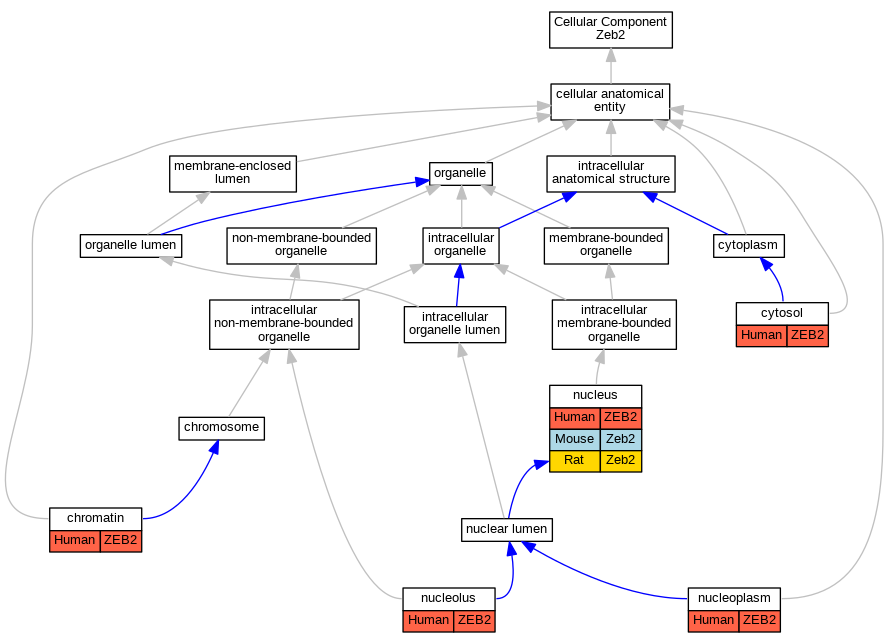
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | Zeb2 | IMP | MGI:MGI:2387499,MGI:MGI:2668659 | Mouse | PMID:25741725 |
| Molecular Function | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | ZEB2 | IDA | | Human | PMID:20516212 |
| Molecular Function | GO:0005515 | protein binding | ZEB2 | IPI | UniProtKB:O94776 | Human | PMID:26028330 |
| Molecular Function | GO:0005515 | protein binding | ZEB2 | IPI | UniProtKB:Q13330 | Human | PMID:26028330 |
| Molecular Function | GO:0005515 | protein binding | ZEB2 | IPI | UniProtKB:Q96KQ7 | Human | PMID:26028330 |
| Molecular Function | GO:0070412 | R-SMAD binding | Zeb2 | IDA | | Mouse | PMID:16115198 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | ZEB2 | IDA | | Human | PMID:20516212 |
| Cellular Component | GO:0000785 | chromatin | ZEB2 | IDA | | Human | PMID:20516212 |
| Cellular Component | GO:0005829 | cytosol | ZEB2 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005730 | nucleolus | ZEB2 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005654 | nucleoplasm | ZEB2 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005634 | nucleus | ZEB2 | IDA | | Human | PMID:24769727 |
| Cellular Component | GO:0005634 | nucleus | Zeb2 | IDA | | Mouse | MGI:MGI:5574413|PMID:24769727 |
| Cellular Component | GO:0005634 | nucleus | Zeb2 | IDA | | Rat | PMID:30336567|RGD:155882532 |
| Cellular Component | GO:0005634 | nucleus | Zeb2 | IDA | | Rat | PMID:34852714|RGD:155882542 |
| Biological Process | GO:0048143 | astrocyte activation | Zeb2 | IDA | | Rat | PMID:34852714|RGD:155882542 |
| Biological Process | GO:0021846 | cell proliferation in forebrain | Zeb2 | IMP | MGI:MGI:2387499,MGI:MGI:2684610 | Mouse | PMID:17644613 |
| Biological Process | GO:0007417 | central nervous system development | Zeb2 | IGI | MGI:MGI:1344313 | Mouse | PMID:16598713 |
| Biological Process | GO:0048668 | collateral sprouting | Zeb2 | IMP | MGI:MGI:2387499,MGI:MGI:2668659 | Mouse | PMID:25741725 |
| Biological Process | GO:0060028 | convergent extension involved in axis elongation | zeb2a | IGI | ZFIN:ZDB-MRPHLNO-080717-4,ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0060028 | convergent extension involved in axis elongation | zeb2a | IGI | ZFIN:ZDB-MRPHLNO-080717-3,ZFIN:ZDB-MRPHLNO-080717-5 | Zfish | PMID:18351671 |
| Biological Process | GO:0060028 | convergent extension involved in axis elongation | zeb2b | IMP | ZFIN:ZDB-MRPHLNO-080717-5 | Zfish | PMID:18351671 |
| Biological Process | GO:0060028 | convergent extension involved in axis elongation | zeb2b | IGI | ZFIN:ZDB-MRPHLNO-080717-3,ZFIN:ZDB-MRPHLNO-080717-5 | Zfish | PMID:18351671 |
| Biological Process | GO:0060028 | convergent extension involved in axis elongation | zeb2b | IMP | ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0060028 | convergent extension involved in axis elongation | zeb2b | IGI | ZFIN:ZDB-MRPHLNO-080717-4,ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0021540 | corpus callosum morphogenesis | Zeb2 | IMP | MGI:MGI:2387499,MGI:MGI:2668659 | Mouse | PMID:25741725 |
| Biological Process | GO:0021957 | corticospinal tract morphogenesis | Zeb2 | IMP | MGI:MGI:2387499,MGI:MGI:2668659 | Mouse | PMID:25741725 |
| Biological Process | GO:0048066 | developmental pigmentation | Zeb2 | IMP | | Mouse | MGI:MGI:5574413|PMID:24769727 |
| Biological Process | GO:0060030 | dorsal convergence | zeb2a | IMP | ZFIN:ZDB-MRPHLNO-080717-3 | Zfish | PMID:18351671 |
| Biological Process | GO:0060030 | dorsal convergence | zeb2a | IGI | ZFIN:ZDB-MRPHLNO-080717-3,ZFIN:ZDB-MRPHLNO-080717-5 | Zfish | PMID:18351671 |
| Biological Process | GO:0060030 | dorsal convergence | zeb2b | IGI | ZFIN:ZDB-MRPHLNO-080717-3,ZFIN:ZDB-MRPHLNO-080717-5 | Zfish | PMID:18351671 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | zeb2a | IGI | ZFIN:ZDB-MRPHLNO-080717-4,ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | zeb2a | IMP | ZFIN:ZDB-MRPHLNO-080717-4 | Zfish | PMID:18351671 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | zeb2b | IMP | ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | zeb2b | IGI | ZFIN:ZDB-MRPHLNO-080717-4,ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0048598 | embryonic morphogenesis | Zeb2 | IGI | MGI:MGI:1344313 | Mouse | PMID:16598713 |
| Biological Process | GO:0043542 | endothelial cell migration | Zeb2 | IMP | | Rat | PMID:31881206|RGD:155882533 |
| Biological Process | GO:0001935 | endothelial cell proliferation | Zeb2 | IMP | | Rat | PMID:31881206|RGD:155882533 |
| Biological Process | GO:0072537 | fibroblast activation | Zeb2 | IDA | | Rat | PMID:30336567|RGD:155882532 |
| Biological Process | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis | zeb2a | IMP | ZFIN:ZDB-MRPHLNO-100521-9 | Zfish | PMID:20034103 |
| Biological Process | GO:0021766 | hippocampus development | Zeb2 | IMP | MGI:MGI:2387499,MGI:MGI:2684610 | Mouse | PMID:17644613 |
| Biological Process | GO:0035622 | intrahepatic bile duct development | zeb2b | IMP | ZFIN:ZDB-MRPHLNO-120222-3 | Zfish | PMID:21336163 |
| Biological Process | GO:0035622 | intrahepatic bile duct development | zeb2b | IMP | ZFIN:ZDB-MRPHLNO-120222-4 | Zfish | PMID:21336163 |
| Biological Process | GO:0061373 | mammillary axonal complex development | Zeb2 | IMP | MGI:MGI:2387499,MGI:MGI:2668659 | Mouse | PMID:25741725 |
| Biological Process | GO:0097324 | melanocyte migration | Zeb2 | IMP | | Mouse | MGI:MGI:5574413|PMID:24769727 |
| Biological Process | GO:0036446 | myofibroblast differentiation | Zeb2 | IEP | | Rat | PMID:24155330|RGD:155882536 |
| Biological Process | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | zeb2a | IMP | ZFIN:ZDB-MRPHLNO-100521-9 | Zfish | PMID:20034103 |
| Biological Process | GO:0010764 | negative regulation of fibroblast migration | Zeb2 | IDA | | Rat | PMID:30336567|RGD:155882532 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ZEB2 | IDA | | Human | PMID:12837246 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ZEB2 | IDA | | Human | PMID:20516212 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ZEB2 | IMP | | Human | PMID:24769727 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | Zeb2 | IMP | | Mouse | MGI:MGI:5574413|PMID:24769727 |
| Biological Process | GO:0001755 | neural crest cell migration | Zeb2 | IMP | MGI:MGI:2387501 | Mouse | PMID:12522767 |
| Biological Process | GO:0001755 | neural crest cell migration | zeb2b | IMP | ZFIN:ZDB-MRPHLNO-130618-1 | Zfish | PMID:23466526 |
| Biological Process | GO:0014029 | neural crest formation | zeb2a | IGI | ZFIN:ZDB-MRPHLNO-080717-4,ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0014029 | neural crest formation | zeb2a | IMP | ZFIN:ZDB-MRPHLNO-080717-4 | Zfish | PMID:18351671 |
| Biological Process | GO:0014029 | neural crest formation | zeb2b | IMP | ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0014029 | neural crest formation | zeb2b | IGI | ZFIN:ZDB-MRPHLNO-080717-4,ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0001843 | neural tube closure | Zeb2 | IMP | MGI:MGI:2387501 | Mouse | PMID:16598713 |
| Biological Process | GO:0050772 | positive regulation of axonogenesis | Zeb2 | IMP | MGI:MGI:2387499,MGI:MGI:2668659 | Mouse | PMID:25741725 |
| Biological Process | GO:0090263 | positive regulation of canonical Wnt signaling pathway | ZEB2 | IMP | | Human | PMID:30592286 |
| Biological Process | GO:0043507 | positive regulation of JUN kinase activity | Zeb2 | IMP | MGI:MGI:2387499,MGI:MGI:2684610 | Mouse | PMID:17644613 |
| Biological Process | GO:1902748 | positive regulation of lens fiber cell differentiation | Zeb2 | IMP | | Mouse | MGI:MGI:3604916|PMID:16162653 |
| Biological Process | GO:0045636 | positive regulation of melanocyte differentiation | Zeb2 | IMP | | Mouse | MGI:MGI:5574413|PMID:24769727 |
| Biological Process | GO:1904330 | positive regulation of myofibroblast contraction | Zeb2 | IDA | | Rat | PMID:30336567|RGD:155882532 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ZEB2 | IMP | | Human | PMID:24769727 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Zeb2 | IMP | | Mouse | MGI:MGI:5574413|PMID:24769727 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Zeb2 | IMP | MGI:MGI:2387499,MGI:MGI:2668659 | Mouse | PMID:25741725 |
| Biological Process | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | ZEB2 | IMP | | Human | PMID:30592286 |
| Biological Process | GO:0030177 | positive regulation of Wnt signaling pathway | Zeb2 | IMP | MGI:MGI:2387499,MGI:MGI:2684610 | Mouse | PMID:17644613 |
| Biological Process | GO:0070269 | pyroptosis | Zeb2 | IDA | | Rat | PMID:34852714|RGD:155882542 |
| Biological Process | GO:1905603 | regulation of blood-brain barrier permeability | Zeb2 | IMP | | Rat | PMID:34334113|RGD:155882538 |
| Biological Process | GO:1903056 | regulation of melanosome organization | Zeb2 | IMP | | Mouse | MGI:MGI:5574413|PMID:24769727 |
| Biological Process | GO:1904520 | regulation of myofibroblast cell apoptotic process | Zeb2 | IMP | | Rat | PMID:34321385|RGD:155882552 |
| Biological Process | GO:0090649 | response to oxygen-glucose deprivation | Zeb2 | IMP | | Rat | PMID:34321385|RGD:155882552 |
| Biological Process | GO:0001756 | somitogenesis | Zeb2 | IMP | MGI:MGI:2387501 | Mouse | PMID:16157277 |
| Biological Process | GO:0001756 | somitogenesis | Zeb2 | IMP | MGI:MGI:2387501 | Mouse | PMID:16598713 |
| Biological Process | GO:0001756 | somitogenesis | zeb2a | IGI | ZFIN:ZDB-MRPHLNO-080717-3,ZFIN:ZDB-MRPHLNO-080717-5 | Zfish | PMID:18351671 |
| Biological Process | GO:0001756 | somitogenesis | zeb2a | IGI | ZFIN:ZDB-MRPHLNO-080717-4,ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0001756 | somitogenesis | zeb2a | IMP | ZFIN:ZDB-MRPHLNO-080717-4 | Zfish | PMID:18351671 |
| Biological Process | GO:0001756 | somitogenesis | zeb2b | IGI | ZFIN:ZDB-MRPHLNO-080717-4,ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0001756 | somitogenesis | zeb2b | IMP | ZFIN:ZDB-MRPHLNO-080717-6 | Zfish | PMID:18351671 |
| Biological Process | GO:0001756 | somitogenesis | zeb2b | IGI | ZFIN:ZDB-MRPHLNO-080717-3,ZFIN:ZDB-MRPHLNO-080717-5 | Zfish | PMID:18351671 |
| Biological Process | GO:0001756 | somitogenesis | zeb2b | IMP | ZFIN:ZDB-MRPHLNO-080717-5 | Zfish | PMID:18351671 |
| Biological Process | GO:0043149 | stress fiber assembly | Zeb2 | IMP | | Rat | PMID:31784544|RGD:155882534 |
 Analysis Tools
Analysis Tools