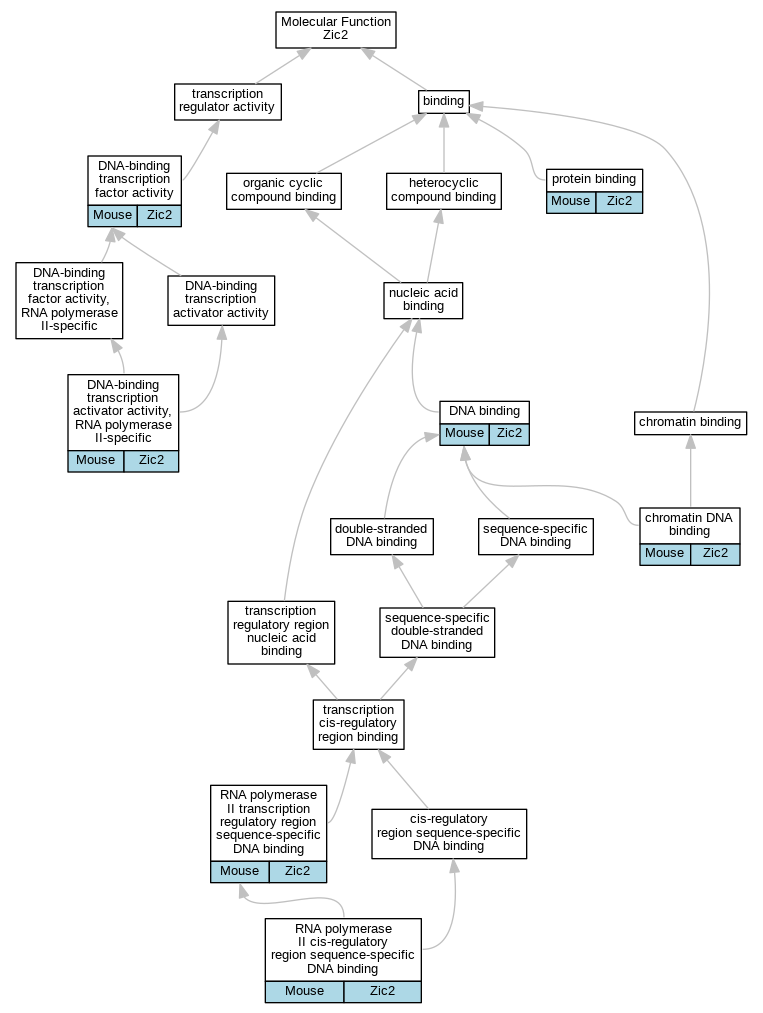
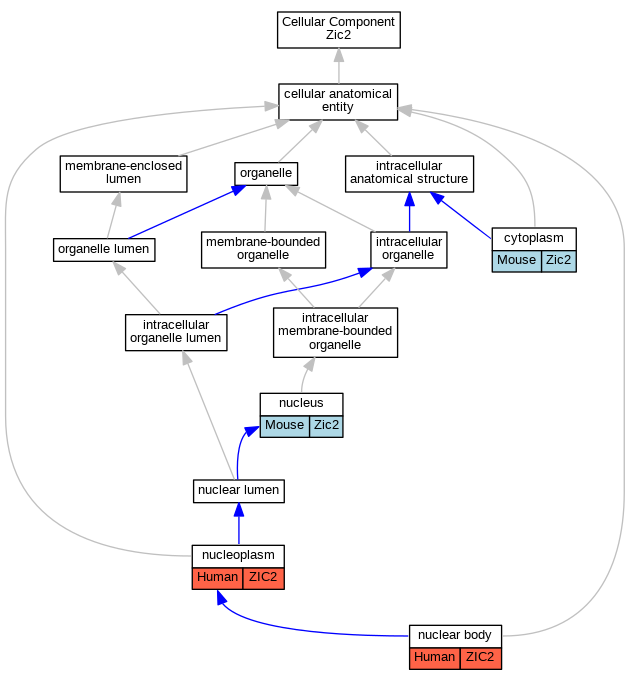
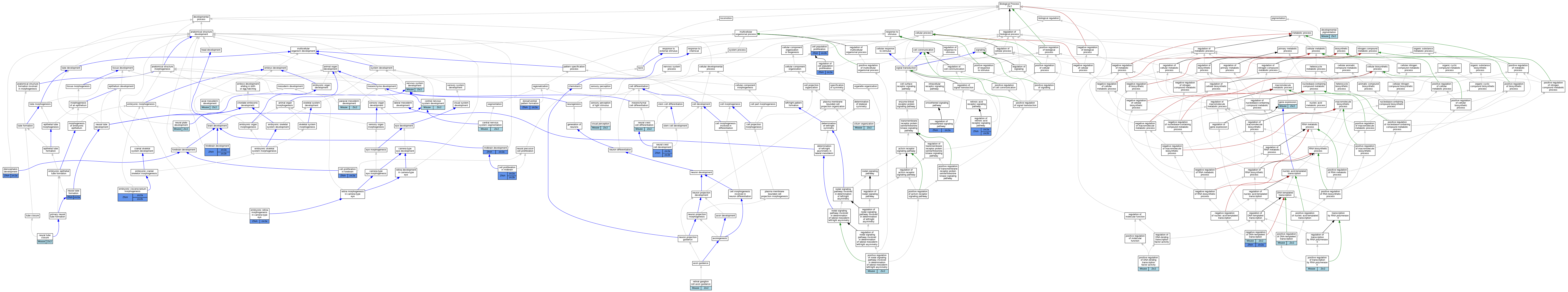
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0031490 | chromatin DNA binding | Zic2 | IDA | | Mouse | MGI:MGI:4833763|PMID:20676059 |
| Molecular Function | GO:0003677 | DNA binding | Zic2 | IDA | | Mouse | MGI:MGI:3771485|PMID:18068128 |
| Molecular Function | GO:0003677 | DNA binding | Zic2 | IDA | | Mouse | PMID:15465018 |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | Zic2 | IDA | | Mouse | MGI:MGI:4942260|PMID:11053430 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | Zic2 | IDA | | Mouse | MGI:MGI:4942260|PMID:11053430 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | Zic2 | IDA | | Mouse | MGI:MGI:3056015|PMID:15465018 |
| Molecular Function | GO:0005515 | protein binding | Zic2 | IPI | UniProtKB:O70133 | Mouse | MGI:MGI:3771485|PMID:18068128 |
| Molecular Function | GO:0005515 | protein binding | Zic2 | IPI | UniProtKB:P10071 | Mouse | MGI:MGI:3688043|PMID:11238441 |
| Molecular Function | GO:0005515 | protein binding | Zic2 | IPI | UniProtKB:Q8BX65 | Mouse | MGI:MGI:4943413|PMID:15207726 |
| Molecular Function | GO:0005515 | protein binding | Zic2 | IPI | UniProtKB:Q8BX65 | Mouse | MGI:MGI:3056015|PMID:15465018 |
| Molecular Function | GO:0005515 | protein binding | Zic2 | IPI | UniProtKB:Q0VGT2 | Mouse | PMID:21211521 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | Zic2 | IDA | | Mouse | MGI:MGI:4942260|PMID:11053430 |
| Molecular Function | GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | Zic2 | IDA | | Mouse | PMID:18298960 |
| Cellular Component | GO:0005737 | cytoplasm | Zic2 | IDA | | Mouse | MGI:MGI:4943413|PMID:15207726 |
| Cellular Component | GO:0005737 | cytoplasm | Zic2 | IDA | | Mouse | MGI:MGI:3056015|PMID:15465018 |
| Cellular Component | GO:0005737 | cytoplasm | Zic2 | IDA | | Mouse | PMID:18755297 |
| Cellular Component | GO:0016604 | nuclear body | ZIC2 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005654 | nucleoplasm | ZIC2 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005634 | nucleus | Zic2 | IDA | | Mouse | MGI:MGI:4943413|PMID:15207726 |
| Cellular Component | GO:0005634 | nucleus | Zic2 | IDA | | Mouse | MGI:MGI:3688043|PMID:11238441 |
| Cellular Component | GO:0005634 | nucleus | Zic2 | IDA | | Mouse | MGI:MGI:3771485|PMID:18068128 |
| Cellular Component | GO:0005634 | nucleus | Zic2 | IDA | | Mouse | PMID:18755297 |
| Cellular Component | GO:0005634 | nucleus | Zic2 | IDA | | Mouse | PMID:12799086 |
| Cellular Component | GO:0005634 | nucleus | Zic2 | IDA | | Mouse | PMID:15465018 |
| Biological Process | GO:0048318 | axial mesoderm development | Zic2 | IGI | MGI:MGI:106676 | Mouse | PMID:17490632 |
| Biological Process | GO:0008283 | cell population proliferation | zic2b | IMP | ZFIN:ZDB-MRPHLNO-180830-11,ZFIN:ZDB-MRPHLNO-180830-10 | Zfish | PMID:29719254 |
| Biological Process | GO:0021846 | cell proliferation in forebrain | zic2a | IMP | ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:18358464 |
| Biological Process | GO:0033278 | cell proliferation in midbrain | zic2b | IMP | | Zfish | PMID:17215296 |
| Biological Process | GO:0033278 | cell proliferation in midbrain | zic2b | IGI | ZFIN:ZDB-GENE-040622-1 | Zfish | PMID:17215296 |
| Biological Process | GO:0033278 | cell proliferation in midbrain | zic2a | IMP | ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:17215296 |
| Biological Process | GO:0007417 | central nervous system development | Zic2 | IGI | MGI:MGI:106683 | Mouse | PMID:11756505 |
| Biological Process | GO:0035283 | central nervous system segmentation | Zic2 | IGI | MGI:MGI:106676 | Mouse | PMID:17490632 |
| Biological Process | GO:0044782 | cilium organization | Zic2 | IMP | MGI:MGI:1862004 | Mouse | PMID:24585447 |
| Biological Process | GO:0048066 | developmental pigmentation | Zic2 | IMP | MGI:MGI:1861983 | Mouse | PMID:10932191 |
| Biological Process | GO:0021536 | diencephalon development | zic2a | IGI | ZFIN:ZDB-GENO-080605-1,ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:18358464 |
| Biological Process | GO:0021536 | diencephalon development | zic2a | IMP | ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:18358464 |
| Biological Process | GO:0021536 | diencephalon development | zic2a | IGI | ZFIN:ZDB-GENO-070406-1,ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:18358464 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | zic2a | IGI | ZFIN:ZDB-GENO-060811-12 | Zfish | PMID:18248235 |
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | zic2a | IMP | | Zfish | PMID:18248235 |
| Biological Process | GO:0060059 | embryonic retina morphogenesis in camera-type eye | zic2a | IMP | ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:17215296 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | zic2a | IMP | ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:23665173 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | zic2b | IMP | ZFIN:ZDB-MRPHLNO-130809-1 | Zfish | PMID:23665173 |
| Biological Process | GO:0010467 | gene expression | Zic2 | IMP | | Mouse | PMID:21211521 |
| Biological Process | GO:0010467 | gene expression | Zic2 | IGI | MGI:MGI:106676 | Mouse | PMID:17490632 |
| Biological Process | GO:0030902 | hindbrain development | zic2a | IGI | ZFIN:ZDB-MRPHLNO-070522-3,ZFIN:ZDB-MRPHLNO-130809-1 | Zfish | PMID:23937294 |
| Biological Process | GO:0030902 | hindbrain development | zic2b | IGI | ZFIN:ZDB-MRPHLNO-070522-3,ZFIN:ZDB-MRPHLNO-130809-1 | Zfish | PMID:23937294 |
| Biological Process | GO:0030901 | midbrain development | zic2a | IMP | ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:17215296 |
| Biological Process | GO:0030901 | midbrain development | zic2a | IGI | ZFIN:ZDB-MRPHLNO-070522-3,ZFIN:ZDB-MRPHLNO-070523-1 | Zfish | PMID:19470582 |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | Zic2 | IDA | | Mouse | MGI:MGI:4942260|PMID:11053430 |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | zic2a | IMP | ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:19855021 |
| Biological Process | GO:0007399 | nervous system development | Zic2 | IMP | MGI:MGI:2156825 | Mouse | PMID:10677508 |
| Biological Process | GO:0014032 | neural crest cell development | zic2a | IMP | ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:23665173 |
| Biological Process | GO:0014032 | neural crest cell development | zic2b | IMP | ZFIN:ZDB-MRPHLNO-130809-1 | Zfish | PMID:23665173 |
| Biological Process | GO:0014033 | neural crest cell differentiation | Zic2 | IMP | MGI:MGI:1862004 | Mouse | PMID:15384171 |
| Biological Process | GO:0001840 | neural plate development | Zic2 | IGI | MGI:MGI:106676 | Mouse | PMID:17490632 |
| Biological Process | GO:0001843 | neural tube closure | Zic2 | IMP | MGI:MGI:1862004 | Mouse | PMID:15384171 |
| Biological Process | GO:0001841 | neural tube formation | zic2a | IGI | ZFIN:ZDB-MRPHLNO-070522-3,ZFIN:ZDB-MRPHLNO-070523-1 | Zfish | PMID:19470582 |
| Biological Process | GO:0048339 | paraxial mesoderm development | Zic2 | IGI | MGI:MGI:106676 | Mouse | PMID:17490632 |
| Biological Process | GO:0051091 | positive regulation of DNA-binding transcription factor activity | Zic2 | IDA | | Mouse | MGI:MGI:4833763|PMID:20676059 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | Zic2 | IDA | | Mouse | MGI:MGI:3056015|PMID:15465018 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | Zic2 | IDA | | Mouse | MGI:MGI:4833763|PMID:20676059 |
| Biological Process | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | Zic2 | IMP | MGI:MGI:1862004 | Mouse | PMID:24585447 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Zic2 | IDA | | Mouse | MGI:MGI:4942260|PMID:11053430 |
| Biological Process | GO:0042127 | regulation of cell population proliferation | zic2a | IMP | ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:17215296 |
| Biological Process | GO:0048385 | regulation of retinoic acid receptor signaling pathway | zic2a | IGI | ZFIN:ZDB-MRPHLNO-070522-3,ZFIN:ZDB-MRPHLNO-130809-1 | Zfish | PMID:23937294 |
| Biological Process | GO:0048385 | regulation of retinoic acid receptor signaling pathway | zic2b | IGI | ZFIN:ZDB-MRPHLNO-070522-3,ZFIN:ZDB-MRPHLNO-130809-1 | Zfish | PMID:23937294 |
| Biological Process | GO:0008589 | regulation of smoothened signaling pathway | zic2a | IMP | ZFIN:ZDB-MRPHLNO-070522-3 | Zfish | PMID:19855021 |
| Biological Process | GO:0031290 | retinal ganglion cell axon guidance | Zic2 | IMP | | Mouse | MGI:MGI:2675195|PMID:13678579 |
| Biological Process | GO:0007601 | visual perception | Zic2 | IMP | | Mouse | MGI:MGI:2675195|PMID:13678579 |
| Biological Process | GO:0007601 | visual perception | Zic2 | IMP | | Mouse | MGI:MGI:4833763|PMID:20676059 |
 Analysis Tools
Analysis Tools