|
Symbol Name ID |
Jmy
junction-mediating and regulatory protein MGI:1913096 |
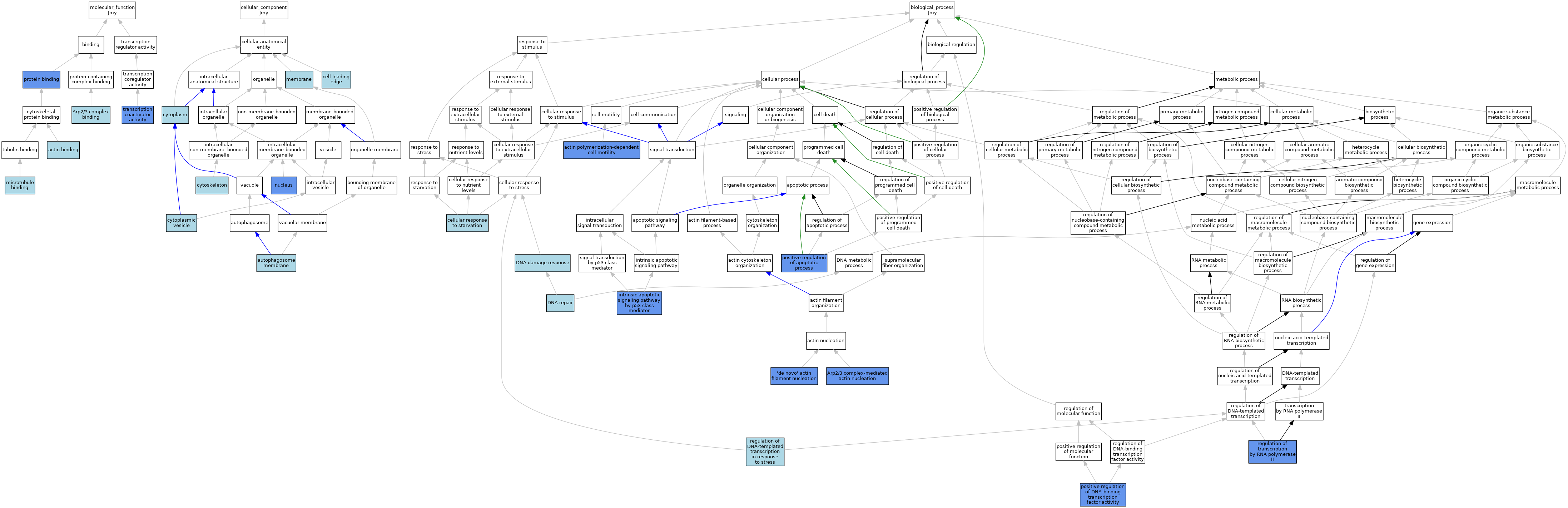
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003779 | actin binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003779 | actin binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0071933 | Arp2/3 complex binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:314962 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:64939 | |||||||||
| Molecular Function | GO:0003713 | transcription coactivator activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003713 | transcription coactivator activity | IDA | J:64939 | |||||||||
| Cellular Component | GO:0000421 | autophagosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031252 | cell leading edge | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:156038 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IC | J:64939 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0070060 | 'de novo' actin filament nucleation | IDA | J:156038 | |||||||||
| Biological Process | GO:0070060 | 'de novo' actin filament nucleation | IBA | J:265628 | |||||||||
| Biological Process | GO:0070358 | actin polymerization-dependent cell motility | IDA | J:156038 | |||||||||
| Biological Process | GO:0034314 | Arp2/3 complex-mediated actin nucleation | IDA | J:156038 | |||||||||
| Biological Process | GO:0034314 | Arp2/3 complex-mediated actin nucleation | IBA | J:265628 | |||||||||
| Biological Process | GO:0009267 | cellular response to starvation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | IEA | J:60000 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IEA | J:60000 | |||||||||
| Biological Process | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | IBA | J:265628 | |||||||||
| Biological Process | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | IDA | J:64939 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | IDA | J:64939 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0051091 | positive regulation of DNA-binding transcription factor activity | IDA | J:64939 | |||||||||
| Biological Process | GO:0051091 | positive regulation of DNA-binding transcription factor activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0043620 | regulation of DNA-templated transcription in response to stress | TAS | J:64939 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IDA | J:64939 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 08/02/2024 MGI 6.24 |

|
|
|
||