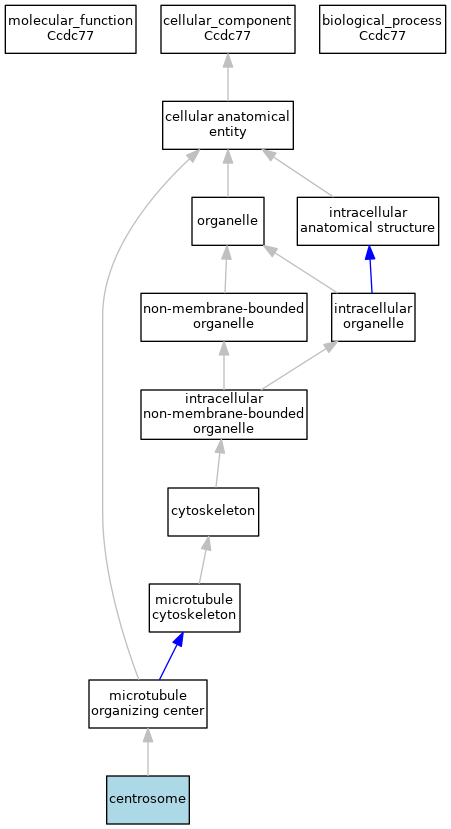
GO Annotations Graph
A
table of the annotations represented in this image is provided below.
- Gray arrows indicate "is_a"; Blue arrows indicate "part_of"; Black arrows indicate "regulates"; Green arrows indicate "positively_regulates"; Red arrows indicate "negatively_regulates"
- Experimental annotations are indicated by darker-colored nodes; other annotations by lighter-colored nodes
- Clicking on any node links to the MGI GO browser and information on all genes annotated to the term or its subterms.
- Graph generated Fri Mar 10 20:53:16 2023.
| Category | GO ID | Classification Term | Evidence | Reference |
| Cellular Component | GO:0005813 | centrosome | ISO | J:164563 |
| Cellular Component | GO:0005813 | centrosome | IBA | J:265628 |
Gene Ontology Evidence Code Abbreviations:
Experimental:
- EXP
- Inferred from experiment
- HMP
- Inferred from high throughput mutant phenotype
- HGI
- Inferred from high throughput genetic interaction
- HDA
- Inferred from high throughput direct assay
- HEP
- Inferred from high throughput expression pattern
- IDA
- Inferred from direct assay
- IEP
- Inferred from expression pattern
- IGI
- Inferred from genetic interaction
- IMP
- Inferred from mutant phenotype
- IPI
- Inferred from physical interaction
Homology:
- IAS
- Inferred from ancestral sequence
- IBA
- Inferred from biological aspect of ancestor
- IBD
- Inferred from biological aspect of descendant
- IKR
- Inferred from key residues
- IMR
- Inferred from missing residues
- IRD
- Inferred from rapid divergence
- ISA
- Inferred from sequence alignment
- ISM
- Inferred from sequence model
- ISO
- Inferred from sequence orthology
- ISS
- Inferred from sequence or structural similarity
Automated:
- IEA
- Inferred from electronic annotation
- RCA
- Reviewed computational analysis
Other:
- IC
- Inferred by curator
- NAS
- Non-traceable author statement
- ND
- No biological data available
- TAS
- Traceable author statement
This site uses cookies.
Some cookies are essential for site operations and others help us analyze use and utility of our web site.
Please refer to our
privacy policy
for more information.
 Analysis Tools
Analysis Tools



