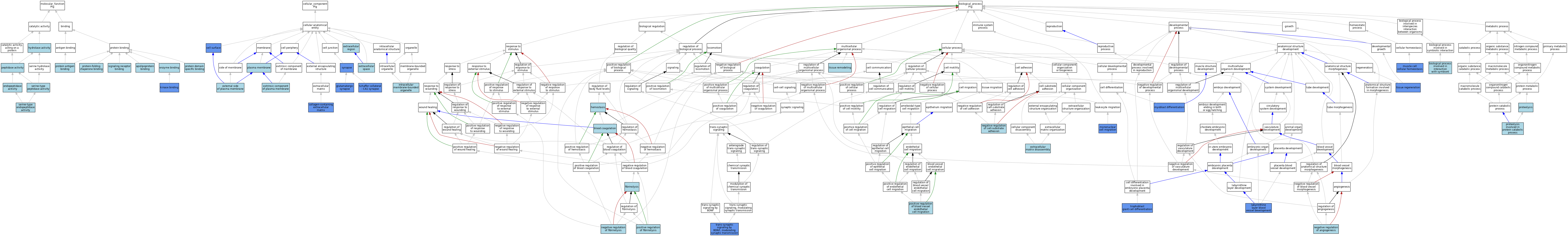
| Category | GO ID | Classification Term | Evidence | Reference |
| Molecular Function | GO:0034185 | apolipoprotein binding | ISO | J:164563 |
| Molecular Function | GO:0004175 | endopeptidase activity | ISO | J:155856 |
| Molecular Function | GO:0004175 | endopeptidase activity | ISO | J:164563 |
| Molecular Function | GO:0004175 | endopeptidase activity | IBA | J:265628 |
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:164563 |
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:164563 |
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:155856 |
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 |
| Molecular Function | GO:0019900 | kinase binding | IPI | J:240586 |
| Molecular Function | GO:0019900 | kinase binding | ISO | J:164563 |
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 |
| Molecular Function | GO:1990405 | protein antigen binding | ISO | J:164563 |
| Molecular Function | GO:0019904 | protein domain specific binding | ISO | J:164563 |
| Molecular Function | GO:0051087 | protein-folding chaperone binding | ISO | J:164563 |
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | ISO | J:164563 |
| Molecular Function | GO:0008236 | serine-type peptidase activity | IEA | J:60000 |
| Molecular Function | GO:0005102 | signaling receptor binding | ISO | J:164563 |
| Molecular Function | GO:0005102 | signaling receptor binding | IBA | J:265628 |
| Cellular Component | GO:0009986 | cell surface | ISO | J:164563 |
| Cellular Component | GO:0009986 | cell surface | IDA | J:149113 |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | HDA | J:265604 |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | HDA | J:265607 |
| Cellular Component | GO:0009897 | external side of plasma membrane | ISO | J:164563 |
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 |
| Cellular Component | GO:0005576 | extracellular region | TAS | Reactome:R-MMU-2533953 |
| Cellular Component | GO:0005615 | extracellular space | ISO | J:155856 |
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 |
| Cellular Component | GO:0005615 | extracellular space | IBA | J:265628 |
| Cellular Component | GO:0019897 | extrinsic component of plasma membrane | IBA | J:265628 |
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:105428 |
| Cellular Component | GO:0098978 | glutamatergic synapse | IEP | J:105428 |
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:105428 |
| Cellular Component | GO:0098978 | glutamatergic synapse | IMP | J:105428 |
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:105428 |
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:155856 |
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 |
| Cellular Component | GO:0098685 | Schaffer collateral - CA1 synapse | IDA | J:105428 |
| Cellular Component | GO:0098685 | Schaffer collateral - CA1 synapse | IDA | J:105428 |
| Cellular Component | GO:0098685 | Schaffer collateral - CA1 synapse | IEP | J:105428 |
| Cellular Component | GO:0098685 | Schaffer collateral - CA1 synapse | IMP | J:105428 |
| Cellular Component | GO:0098685 | Schaffer collateral - CA1 synapse | IDA | J:105428 |
| Cellular Component | GO:0045202 | synapse | IMP | J:105428 |
| Biological Process | GO:0051702 | biological process involved in interaction with symbiont | ISO | J:164563 |
| Biological Process | GO:0007596 | blood coagulation | ISO | J:164563 |
| Biological Process | GO:0022617 | extracellular matrix disassembly | ISO | J:164563 |
| Biological Process | GO:0042730 | fibrinolysis | ISO | J:164563 |
| Biological Process | GO:0007599 | hemostasis | IEA | J:60000 |
| Biological Process | GO:0060716 | labyrinthine layer blood vessel development | IMP | J:84595 |
| Biological Process | GO:0071674 | mononuclear cell migration | IMP | J:149113 |
| Biological Process | GO:0046716 | muscle cell cellular homeostasis | IMP | J:76082 |
| Biological Process | GO:0045445 | myoblast differentiation | IMP | J:76082 |
| Biological Process | GO:0016525 | negative regulation of angiogenesis | TAS | J:78784 |
| Biological Process | GO:0010812 | negative regulation of cell-substrate adhesion | ISO | J:164563 |
| Biological Process | GO:0051918 | negative regulation of fibrinolysis | ISO | J:164563 |
| Biological Process | GO:0043536 | positive regulation of blood vessel endothelial cell migration | ISO | J:164563 |
| Biological Process | GO:0051919 | positive regulation of fibrinolysis | ISO | J:164563 |
| Biological Process | GO:0006508 | proteolysis | ISO | J:164563 |
| Biological Process | GO:0006508 | proteolysis | IBA | J:265628 |
| Biological Process | GO:0051603 | proteolysis involved in protein catabolic process | ISO | J:155856 |
| Biological Process | GO:0042246 | tissue regeneration | IMP | J:76082 |
| Biological Process | GO:0048771 | tissue remodeling | IEA | J:60000 |
| Biological Process | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | IDA | J:105428 |
| Biological Process | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | IMP | J:105428 |
| Biological Process | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | IEP | J:105428 |
| Biological Process | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | IDA | J:105428 |
| Biological Process | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | IMP | J:105428 |
| Biological Process | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | IEP | J:105428 |
| Biological Process | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | IDA | J:105428 |
| Biological Process | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | IDA | J:105428 |
| Biological Process | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | IDA | J:105428 |
| Biological Process | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | IDA | J:105428 |
| Biological Process | GO:0060707 | trophoblast giant cell differentiation | IMP | J:84595 |
Gene Ontology Evidence Code Abbreviations:
Experimental:
- EXP
- Inferred from experiment
- HMP
- Inferred from high throughput mutant phenotype
- HGI
- Inferred from high throughput genetic interaction
- HDA
- Inferred from high throughput direct assay
- HEP
- Inferred from high throughput expression pattern
- IDA
- Inferred from direct assay
- IEP
- Inferred from expression pattern
- IGI
- Inferred from genetic interaction
- IMP
- Inferred from mutant phenotype
- IPI
- Inferred from physical interaction
Homology:
- IAS
- Inferred from ancestral sequence
- IBA
- Inferred from biological aspect of ancestor
- IBD
- Inferred from biological aspect of descendant
- IKR
- Inferred from key residues
- IMR
- Inferred from missing residues
- IRD
- Inferred from rapid divergence
- ISA
- Inferred from sequence alignment
- ISM
- Inferred from sequence model
- ISO
- Inferred from sequence orthology
- ISS
- Inferred from sequence or structural similarity
Automated:
- IEA
- Inferred from electronic annotation
- RCA
- Reviewed computational analysis
Other:
- IC
- Inferred by curator
- NAS
- Non-traceable author statement
- ND
- No biological data available
- TAS
- Traceable author statement
This site uses cookies.
Some cookies are essential for site operations and others help us analyze use and utility of our web site.
Please refer to our
privacy policy
for more information.
 Analysis Tools
Analysis Tools