Humans and mice homozygous for recessive mutations in the FOXN1 (forkhead box N1) gene display common phenotypes:
|
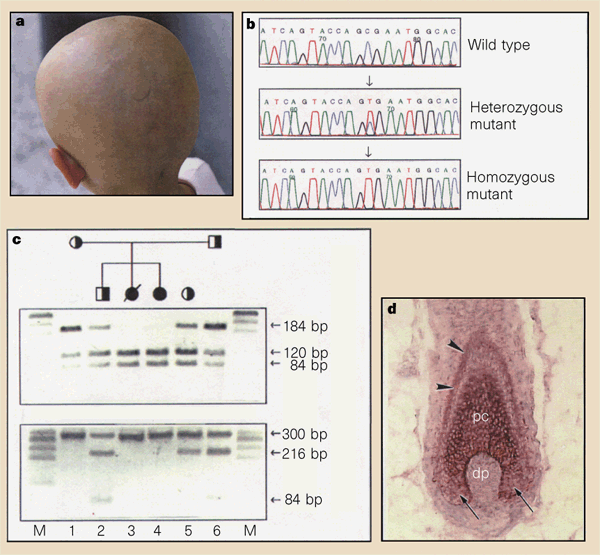
Figure 1 (click to zoom): Molecular analysis of the human nude phenotype. (a) A five-year-old child with congenital alopecia and T-cell immunodeficiency, corrected by bone marrow transplant. (b) Sequence analysis of a nonsense mutation in exon 5 of the FOXN1 gene. Arrow indicates the R255X mutation, a C-to-T transition, causing substitution of an arginine residue by a nonsense mutation. (c) Restriction-enzyme digestion confirming the mutation. (d) FOXN1 mRNA expression in normal human scalp skin. In the hair bulb, FOXN1 mRNA is localized to the differentiating cells of the hair follicle precortex (pc) and the innermost cell layer of the outer root sheath (arrowheads); the dermal papilla (dp) fibroblasts and hair matrix below the level of Auber (small arrows) remain negative for FOXN1 mRNA. From: Frank J, Pignata C, Panteleyev AA, et al. 1999. Exposing the human nude phenotype. Nature 398: 473-474. PubMed: 10206641 Displayed with permission of the Nature Publishing Group. |
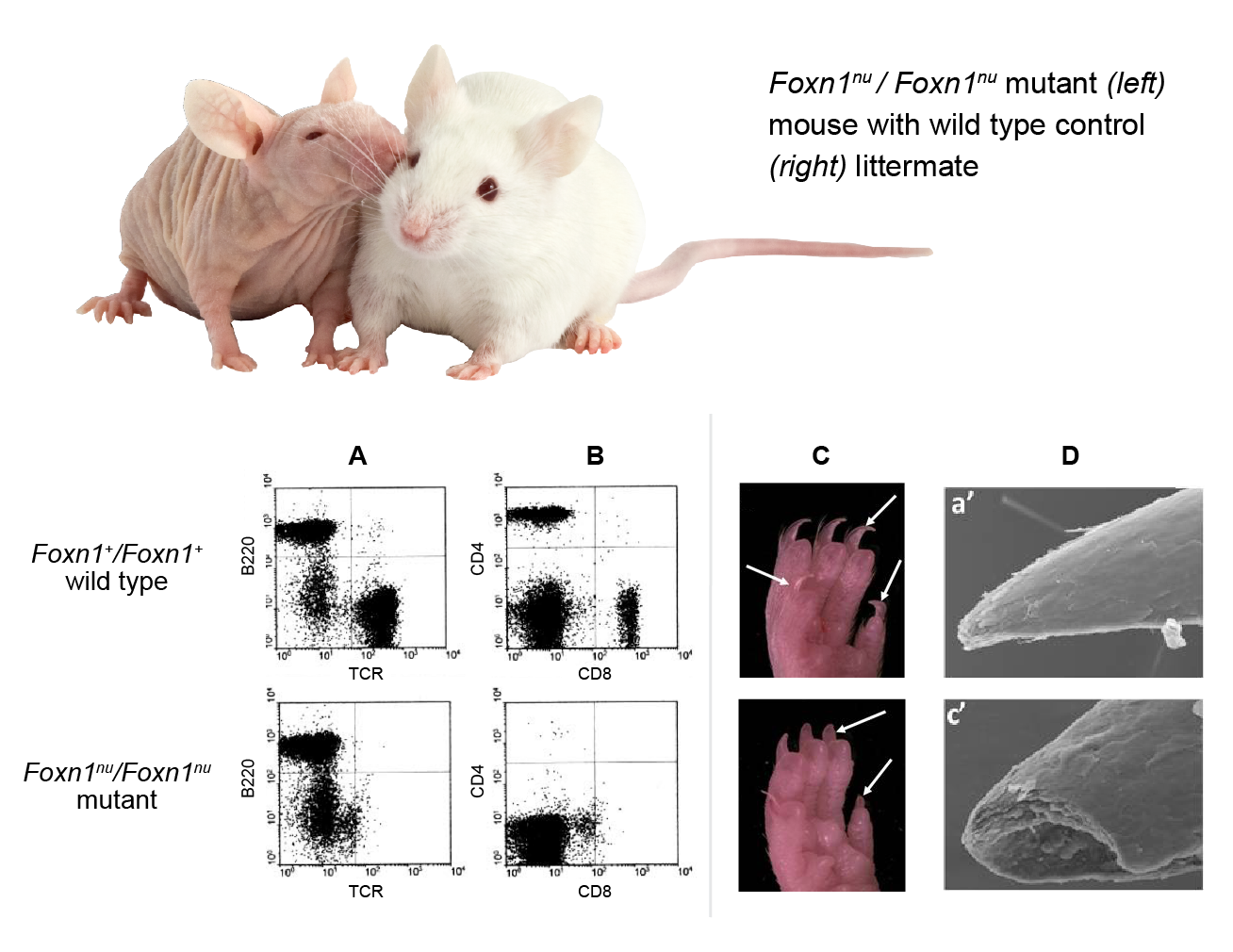 |
Figure 2 (click to zoom): Phenotypic comparison of normal wild type and immunodeficient mutant Foxn1nu/Foxn1nu mouse strains. The ‘nude’ (Foxn1nu) mutation is a single base-pair (G) deletion in exon 3, resulting in a frameshift mutation and termination at a TGA stop codon in exon 6. Additional spontaneous and genetically engineered mouse Foxn1 mutations differing in their sequence changes are known. See Table 1 below. lower panel: FACS profiling of peripheral blood leukocytes shows absence of T-lymphocyte markers in the mutant mouse when stained for (A) the T-cell receptor (TCR) and B-lymphoid marker, B220 or (B) T-cell activation co-receptors CD4 and CD8. Nail morphology in wild-type and mutant mice. Shown are (C) hind-paw photos and (D) scanning EM of nails. Nail plates of normal adults are smooth, hard, and have sharp tips while mutant nails are broken and have blunt ends. A & B figures from: Cunliffe VT, Furley AJ, Keenan D. 2002. Complete rescue of the nude mutant phenotype by a wild-type Foxn1 transgene. Mamm Genome 13:245-252. PubMed: 12016512. Displayed with permission of Mammalian Genome, Springer. C & D figures from: Cai J, Ma L. 2011. Msx2 and Foxn1 regulate nail homeostasis. Genesis 49:449-459. PubMed: 21387539 . Displayed with permission of Genesis, Wiley-Liss, Inc. |
For an overview of mouse Foxn1 mutant phenotypes, you can enter Foxn1 in the “ Search by genes” box on the Human-Mouse: Disease Connection.
Recent articles:
| Allele | Allele name | Mutation type | Mutation detail | Reference |
|---|---|---|---|---|
| Foxn1nu | nude | spontaneous | single base-pair deletion in exon 3 | PMID:7969402 |
| Foxn1nu-2J | nude 2 Jackson | spontaneous | seven base-pair deletion in exon 3, resulting in a frameshift and early termination | https://www.jax.org/strain/016195 |
| Foxn1nu-str | nude streaker | spontaneous | no information | PMID:16068138 |
| Foxn1nu-Y | nude Yuriovo | spontaneous | missense point mutation in exon 7 | PMID:10767081 |
| Foxn1nu-Stl | nude St. Louis | spontaneous | two base-air insertion in exon 7 | PMID:10878619 |
| Foxn1nu-Bc | nude British Columbia | spontaneous | intragenic insertion of an early transposon sequence between exons 1b and 2 | PMID:10348635 |
| Foxn1tw | traveling wave | spontaneous | abnormal splicing | PMID:12893877 |
| Foxn1tm1Tbo | targeted mutation 1, Thomas Boehm | targeted | insertion of an IRES-beta galactosidase-neomycin cassette into exon 3 | PMID:8629026 |
| Foxn1tm1Nrm | targeted mutation 1, Nancy R Manley | targeted | insertion of a GFP-encoding sequence into exon 3, results in aberrant splicing | PMID:14528302 |
| Foxn1tm1.1Cbln | targeted mutation 1.1 Clare Blackburn | targeted | a revertible hypomorph was created by inserting a loxP cassette including a splice acceptor, SV40T antigen, IRES-EGFP/neo and CMAX transcription pause into exon 1b | PMID:22072979 |
Humans and mice heterozygous for mutations in the COL4A1 (collagen, type IV, alpha 1) gene display common phenotypes:
For an overview of mouse Col4a1 mutant phenotypes, you can enter Col4a1 in the “Search by genes” box on the Human-Mouse: Disease Connection.
|
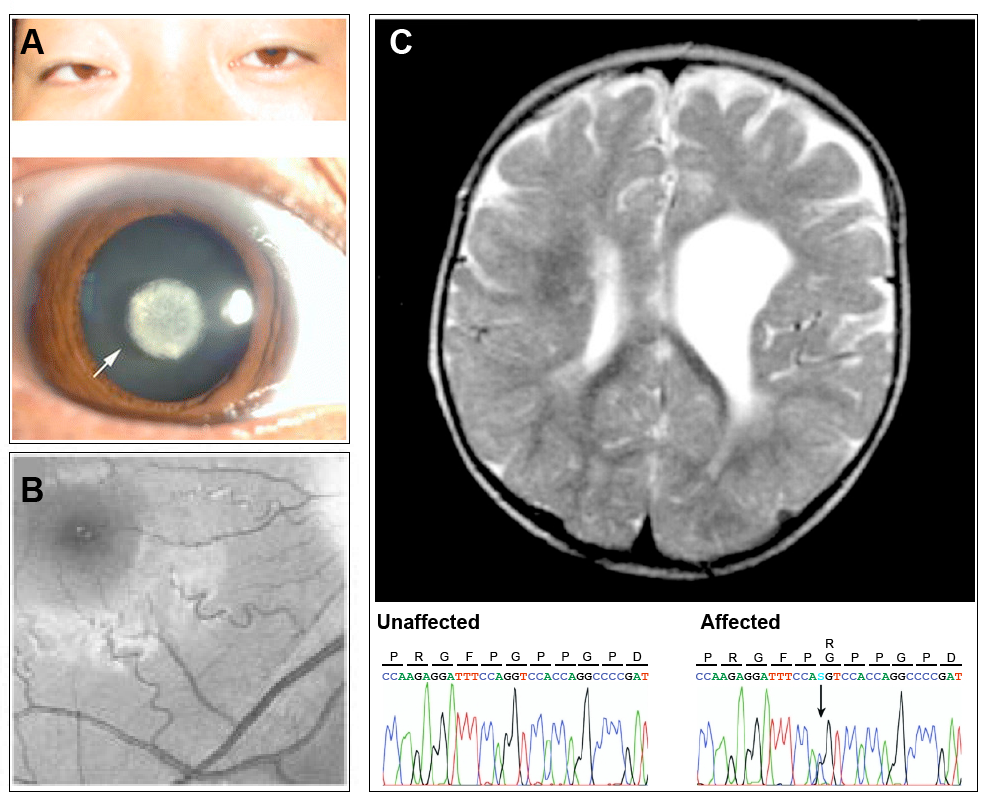
Figure 1 (click to zoom): Characterization of the human COL4A1 mutant phenotype. (A) upper: slit lamp photograph of the dilated eye in an affected patient and lower: detail of nuclear cataract. (B) Retinal arterial tortuosity of medium and small arterioles in the right eye of a 20 year old patient carrying a heterozygous missense mutation in COL4A1. (C) Brain magnetic resonance imaging (MRI) of a 1 year old patient showing porencephalic enlargement of the left ventricle in the frontal area. Sequencing results for this patient below showing a heterozygous G-to-C DNA change leading to a G1423R amino acid substitution. Panel A from: Xia XY, et al., 2014. A novel COL4A1 gene mutation results in autosomal dominant non-syndromic congenital cataract in a Chinese family. BMC Medical Genetics; 15:97. PubMed: 25124159. Reproduced under the terms of the Creative Commons Attribution License. Panel B from: Gould DB et al., 2006. Role of COL4A1 in Small-Vessel Disease and Hemorrhagic Stroke. New England Journal of Medicine; 354:1489-1496 Pubmed: 16598045. Reproduced with permission. Panel C from: Breedveld G., 2006. Novel mutations in three families confirm a major role of COL4A1 in hereditary porencephaly. J Med Genet.; 43(6): 490–495. Pubmed: 16107487. Reproduced with permission. |
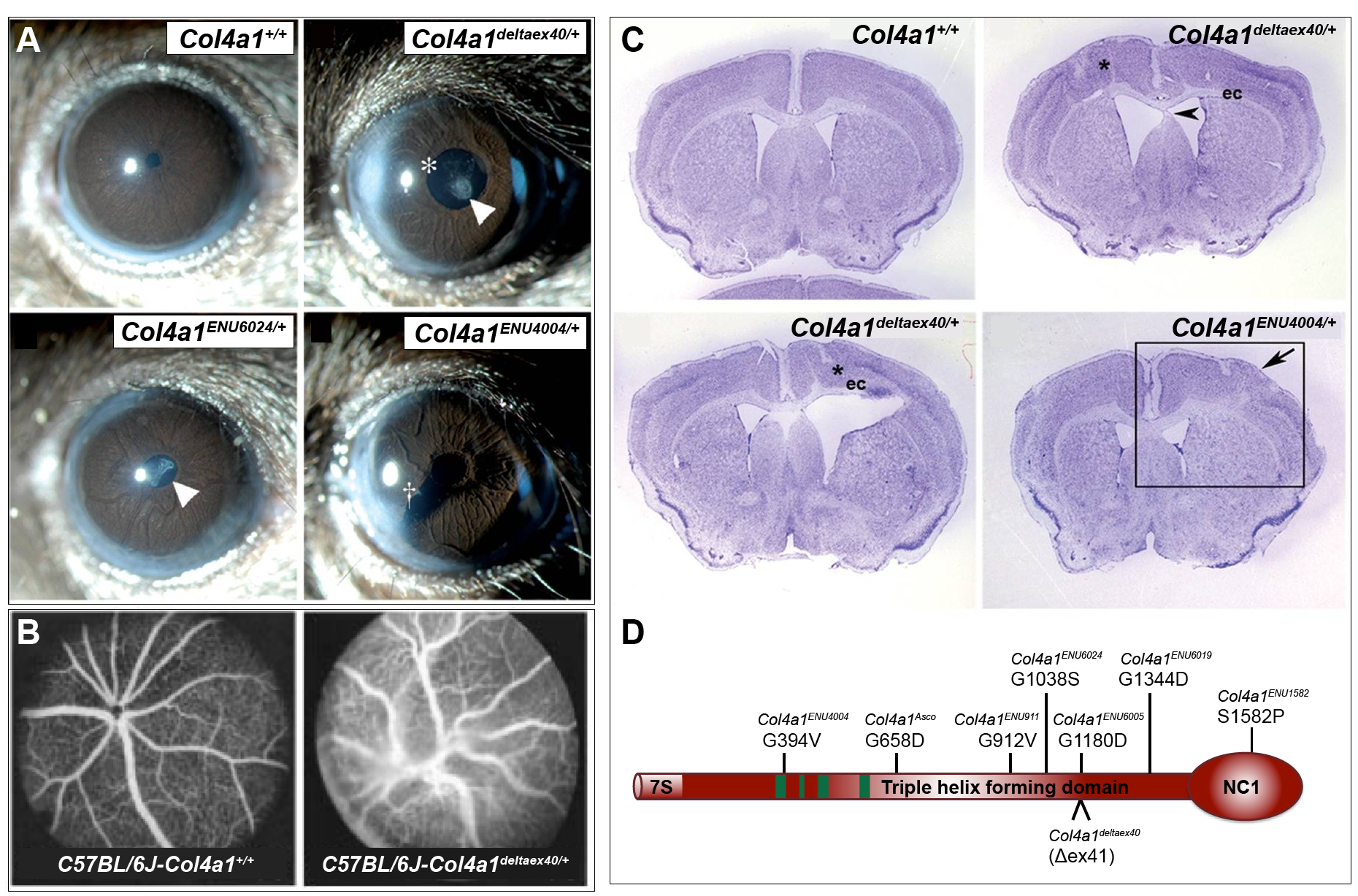 |
Figure 2 (click to zoom): Phenotypic comparison of normal wild type and Col4a1 mutant mouse strains. Images have been re–labeled from originals to reflect official allele nomenclature. (A) Representative images from slit lamp examination of control (Col4a1+/+) and mutant (Col4a1+/mut) eyes showing ocular anterior segment dysgenesis in mutant mice, including open pupil (asterisk), enlarged and torturous iris vasculature, cataracts (arrowhead), and iridocorneal adhesion (cross). (B) Fluorescein angiography in control mice (left) and Col4a1-mutant mice (Col4a1deltaex40, right) on a C57BL/6J genetic background revealed abnormal patterning of the retinal vasculature in mutant mice alone. Mutant retinal vessels were highly tortuous, with more frequent branching than control vessels. (C) Representative images of coronal brain sections from Col4a1+/+ and Col4a1+/mut mice stained with cresyl violet show diverse cortical defects in mutant mice. Defects observed include molecular layer heterotrophia (boxed area in bottom right), focal heterotrophia (asterisks) resembling extensive foliation, enlarged ventricles and white matter defects. (D) Schematic representation of the COL4A1 protein along with phenotypic alleles characterized in Kuo (2014). COL4A1 contains a long, triple–helix–forming, collagenous domain flanked by a short 7S domain at the amino terminus and a globular, non-collagenous (NC1) domain at the carboxy terminus. Green boxes indicate putative integrin–binding sites. Panel A, C & D from: Kuo DS et al., 2014. Allelic heterogeneity contributes to variability in ocular dysgenesis, myopathy and brain malformations caused by Col4a1 and Col4a2 mutations. Hum Mol Genet.; 23(7): 1709–1722. Pubmed: 24203695. Reproduced under the terms of the Creative Common Attribution License. Panel B from: Gould DB et al., 2007. Role of COL4A1 in Small–Vessel Disease and Hemorrhagic Stroke. New England Journal of Medicine; 354:1489-1496 Pubmed: 16598045. Reproduced with permission. |
Additional recent articles and reviews:
| Allele | Allele name | Mutation type | Mutation detail | Original Reference |
|---|---|---|---|---|
| Col4a1Acso | anterior capsular and suture opacity | chemically induced (other) | G to A nucleotide mutation at position 2104 resulting in a G658B amino acid substitution | J:163225 |
| Col4a1Bru | bruised | chemically induced (ENU) | Point mutation in exon 26 resulted in a G627W amino acid substitution | J:102749 |
| Col4a1D456 | radiation induced mutation D456 | radiation induced | G to A nucleotide mutation at position 2991 resulting in a G954R amino acid substitution | J:163225 |
| Col4a1delta40 | delta exon 40 | chemically induced (other) | A mutation in the splice acceptor site of exon 40‡; resulted in the direct
splicing of exon 39 to exon 41. ‡The skipped exon was re–numbered as exon 41 in genome build GRCm38/mm10 [Ref] |
J:98572 |
| Col4a1ENU911 | ENU mutation 911 | chemically induced (ENU) | G to T nucleotide mutation at position 2866 resulting in a G912V amino acid substitution | J:163225 |
| Col4a1ENU4004 | ENU mutation 4004 | chemically induced (ENU) | G to T nucleotide mutation at position 1312 resulting in a G394V amino acid substitution | J:163225 |
| Col4a1ENU6005 | ENU mutation 6005 | chemically induced (ENU) | G to A nucleotide mutation at position 3670 resulting in a G1180D amino acid substitution | J:163225 |
| Col4a1ENU6009 | ENU mutation 6009 | chemically induced (ENU) | T to C nucleotide mutation at position 4875 resulting in an S1582P amino acid substitution | J:163225 |
| Col4a1ENU6019 | ENU mutation 6019 | chemically induced (ENU) | G to A nucleotide mutation at position 4162 resulting in a G1344D amino acid substitution | J:163225 |
| Col4a1ENU6024 | ENU mutation 6024 | chemically induced (ENU) | G to A nucleotide mutation at position 3243 resulting in a G1038S amino acid substitution | J:163225 |
| Col4a1F247 | radiation induced mutation F247 | radiation induced | G to C nucleotide mutation at position 4152 resulting in a G1341R amino acid substitution | J:163225 |
| Col4a1Raw | retinal arteriolar wiring | chemically induced (ENU) | Point mutation in exon 34 resulted in a K950E amino acid substitution | J:102749 |
| Col4a1Svc | small with vacuolar cataract | chemically induced (ENU) | Point mutation in exon 37 resulted in a G1064D amino acid substitution | J:102749 |
| Del(8Col4a1-Col4a2)1Epo | deletion, Chr8, Ernst Poschl 1 | Targeted (Null/knockout), Intergenic deletion | Exon 1 of Col4a1 and exons 1-3 of Col4a2 were replaced with a neo cassette | J:89190 |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 01/06/2026 MGI 6.24 |

|
|
|
||