| Column |
Type |
Size |
Nulls |
Auto |
Default |
Children |
Parents |
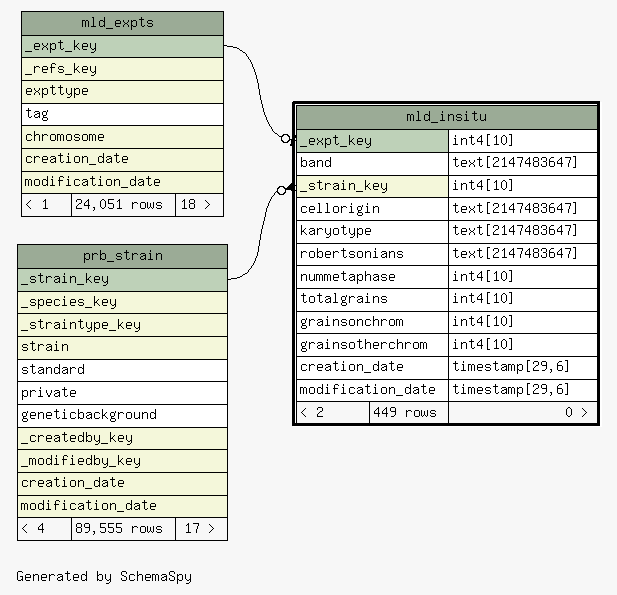
| _expt_key |
int4 |
10 |
|
|
|
|
| mld_expts._expt_key
|
mld_insitu__expt_key_fkey C |
|
| band |
text |
2147483647 |
√ |
|
null |
|
|
| _strain_key |
int4 |
10 |
|
|
|
|
| prb_strain._strain_key
|
mld_insitu__strain_key_fkey R |
|
| cellorigin |
text |
2147483647 |
√ |
|
null |
|
|
| karyotype |
text |
2147483647 |
√ |
|
null |
|
|
| robertsonians |
text |
2147483647 |
√ |
|
null |
|
|
| nummetaphase |
int4 |
10 |
√ |
|
null |
|
|
| totalgrains |
int4 |
10 |
√ |
|
null |
|
|
| grainsonchrom |
int4 |
10 |
√ |
|
null |
|
|
| grainsotherchrom |
int4 |
10 |
√ |
|
null |
|
|
| creation_date |
timestamp |
29,6 |
|
|
now() |
|
|
| modification_date |
timestamp |
29,6 |
|
|
now() |
|
|
Table contained 449 rows at Tue Feb 17 06:11 EST 2026
|